 |
Rosetta
2020.37
|
All Classes Namespaces Files Functions Variables Typedefs Enumerations Enumerator Friends Macros Pages
 |
Rosetta
2020.37
|
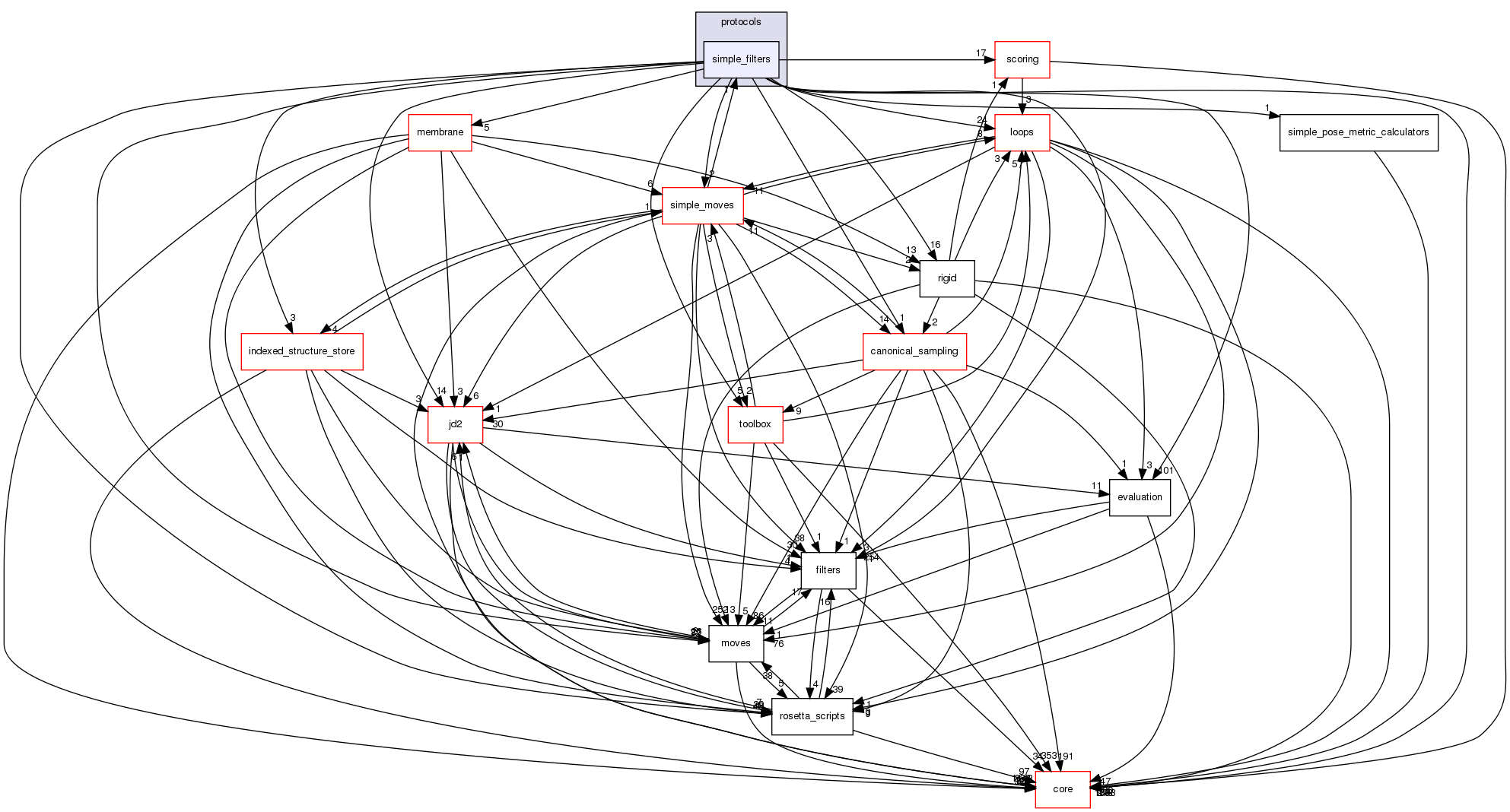
Files | |
| file | AbinitioBaseFilter.cc |
| file | AbinitioBaseFilter.hh |
| header file for AbinitioBaseFilter.cc | |
| file | AngleToVectorFilter.cc |
| file | AngleToVectorFilter.fwd.hh |
| file | AngleToVectorFilter.hh |
| file | AngleToVectorFilterCreator.hh |
| FilterCreators for the AngleToVectorFilter. | |
| file | AtomicContactFilter.cc |
| file | AtomicContactFilter.hh |
| file | AtomicContactFilterCreator.hh |
| FilterCreator for the AtomicContactFilter. | |
| file | AtomicDistanceFilter.cc |
| file | AtomicDistanceFilter.hh |
| Filter for looking at specific atom distances. | |
| file | AtomicDistanceFilterCreator.hh |
| FilterCreator for the AtomicDistanceFilter. | |
| file | AveragePathLengthFilter.cc |
| Filter on the average covalent path length between residues, including disulfides. | |
| file | AveragePathLengthFilter.fwd.hh |
| Filter on the average covalent path length between residues, including disulfides. | |
| file | AveragePathLengthFilter.hh |
| Filter on the average covalent path length between residues, including disulfides. | |
| file | AveragePathLengthFilterCreator.hh |
| FilterCreators for the AveragePathLengthFilter. | |
| file | BatchNrEvaluator.cc |
| file | BatchNrEvaluator.hh |
| file | BuriedSurfaceAreaFilter.cc |
| Calculates buried surface area (exposed surface area minus total surface area, on a per-residue basis). Accepts a residue selector to allow buried subsets to be considered. | |
| file | BuriedSurfaceAreaFilter.fwd.hh |
| Forward declarations and owning pointer typedefs for the BuriedSurfaceAreaFilter. | |
| file | BuriedSurfaceAreaFilter.hh |
| Headers for the BuriedSurfaceAreaFilter. | |
| file | BuriedSurfaceAreaFilterCreator.hh |
| Creator for the BuriedSurfaceAreaFilter. | |
| file | BuriedUnsatHbondFilter.cc |
| filter to report buried polar atoms that are not satisfied by hydrogen bonds | |
| file | BuriedUnsatHbondFilter.fwd.hh |
| file | BuriedUnsatHbondFilter.hh |
| filter to report buried polar atoms that are not satisfied by hydrogen bonds | |
| file | BuriedUnsatHbondFilterCreator.hh |
| FilterCreators for the BuriedUnsatHbondFilterCreator. | |
| file | CamShiftEvaluator.cc |
| file | CamShiftEvaluator.hh |
| file | CamShiftEvaluatorCreator.cc |
| file | CamShiftEvaluatorCreator.hh |
| Header for CamShiftEvaluatorCreator. | |
| file | ChainBreakFilter.cc |
| file | ChainBreakFilter.fwd.hh |
| file | ChainBreakFilter.hh |
| file | ChainBreakFilterCreator.hh |
| FilterCreators for the ChainBreakFilter. | |
| file | ChainCountFilter.cc |
| file | ChainCountFilter.fwd.hh |
| FilterCreator for the ChainCountFilter. | |
| file | ChainCountFilter.hh |
| Count the amount of chains in a pose. | |
| file | ChainCountFilterCreator.hh |
| FilterCreator for the ChainCountFilter. | |
| file | ChiWellRmsdEvaluator.cc |
| file | ChiWellRmsdEvaluator.hh |
| file | ChiWellRmsdEvaluatorCreator.cc |
| file | ChiWellRmsdEvaluatorCreator.hh |
| Header for ChiWellRmsdEvaluatorCreator. | |
| file | COFilter.cc |
| runs reject or accept filters on pose | |
| file | COFilter.hh |
| file | ContactMapEvaluator.cc |
| file | ContactMapEvaluator.hh |
| file | ContactMapEvaluatorCreator.cc |
| file | ContactMapEvaluatorCreator.hh |
| Header for ContactMapEvaluatorCreator. | |
| file | ContactMolecularSurfaceFilter.cc |
| Filter structures by weighted contact molecular surface area. | |
| file | ContactMolecularSurfaceFilter.fwd.hh |
| Filter structures by weighted contact molecular surface area. | |
| file | ContactMolecularSurfaceFilter.hh |
| header file for ContactMolecularSurfaceFilter class | |
| file | ContactMolecularSurfaceFilterCreator.hh |
| FilterCreator for the ContactMolecularSurfaceFilter. | |
| file | DeltaFilter.cc |
| file | DeltaFilter.fwd.hh |
| forward declaration for DeltaFilter | |
| file | DeltaFilter.hh |
| Reports the average degree of connectivity of interface residues. | |
| file | DeltaFilterCreator.hh |
| FilterCreators for the DeltaFilter. | |
| file | DisulfideEntropyFilter.cc |
| Filter on the entropic effect of disulfide linkage. | |
| file | DisulfideEntropyFilter.fwd.hh |
| Filter on the entropic effect of disulfide linkage. | |
| file | DisulfideEntropyFilter.hh |
| Filter on the entropic effect of disulfide linkage. | |
| file | DisulfideEntropyFilterCreator.hh |
| FilterCreators for the DisulfideEntropyFilter. | |
| file | DomainInterfaceFilter.cc |
| Implementation of DomainInterfaceFilter which checks whether a defined region of a protein is part of an interface with a pre-defined other domain of the protein. | |
| file | DomainInterfaceFilter.fwd.hh |
| forward declaration for DomainInterfaceFilter which checks if a query region is part of an an interface with a pre-defined other domain of a protein. | |
| file | DomainInterfaceFilter.hh |
| header file for DomainInterfaceFilter which checks whether a defined region of a protein is buried in an interface by evaluating the number of non-water neighbors of all residues | |
| file | EnergyPerResidueFilter.cc |
| file | EnergyPerResidueFilter.fwd.hh |
| file | EnergyPerResidueFilter.hh |
| definition of filter class EnergyPerResidueFilter. | |
| file | EnergyPerResidueFilterCreator.hh |
| FilterCreators for the EnergyPerResidueFilterCreator. | |
| file | EvaluatedTrialMover.cc |
| file | EvaluatedTrialMover.hh |
| file | ExpiryFilter.cc |
| file | ExpiryFilter.fwd.hh |
| file | ExpiryFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | ExpiryFilterCreator.hh |
| FilterCreators for the ExpiryFilter. | |
| file | ExternalEvaluator.cc |
| file | ExternalEvaluator.hh |
| file | ExtraScoreEvaluatorCreator.cc |
| file | ExtraScoreEvaluatorCreator.hh |
| Header for ExtraScoreEvaluatorCreator. | |
| file | FileExistFilter.cc |
| file | FileExistFilter.fwd.hh |
| file | FileExistFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | FileExistFilterCreator.hh |
| FilterCreators for the FileExistFilter. | |
| file | FileRemoveFilter.cc |
| file | FileRemoveFilter.fwd.hh |
| file | FileRemoveFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | FileRemoveFilterCreator.hh |
| FilterCreators for the FileRemoveFilter. | |
| file | HelixHelixAngleFilter.cc |
| HelixHelixAngleFilter computes wither the crossing angle or distance between two TMs. | |
| file | HelixHelixAngleFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | HelixHelixAngleFilter.hh |
| HelixHelixAngleFilter computes wither the crossing angle or distance between two TMs. | |
| file | HelixHelixAngleFilterCreator.hh |
| FilterCreators for the HelixHelixAngleFilterCreator. | |
| file | HolesFilter.cc |
| file | HolesFilter.fwd.hh |
| filter structures by will's hole score | |
| file | HolesFilter.hh |
| header file for HolesFilter class. | |
| file | HolesFilterCreator.hh |
| FilterCreator for the HolesFilter. | |
| file | InterfaceHbondsFilter.cc |
| Counts the number of hydrogen bonds across an interface defined by a jump, the salt. | |
| file | InterfaceHbondsFilter.fwd.hh |
| forward declaration for InterfaceHbondsFilter | |
| file | InterfaceHbondsFilter.hh |
| Counts the number of cross interface hydrogen bonds. | |
| file | InterfaceHbondsFilterCreator.hh |
| FilterCreators for the InterfaceHbondsFilter. | |
| file | InterfaceHydrophobicResidueContactsFilter.cc |
| Counts the number of hydrophobic residues on the target that have at least a certain hydrophobic score. | |
| file | InterfaceHydrophobicResidueContactsFilter.fwd.hh |
| forward declaration for InterfaceHydrophobicResidueContactsFilter | |
| file | InterfaceHydrophobicResidueContactsFilter.hh |
| Counts the number of hydrophobic residues on the target that have at least a certain hydrophobic score. | |
| file | InterfaceHydrophobicResidueContactsFilterCreator.hh |
| FilterCreators for the InterfaceHydrophobicResidueContactsFilter. | |
| file | InterfaceSasaFilter.cc |
| file | InterfaceSasaFilter.fwd.hh |
| file | InterfaceSasaFilter.hh |
| definition of filter class InterfaceSasaFilter. | |
| file | InterfaceSasaFilterCreator.hh |
| FilterCreators for the InterfaceSasaFilterCreator. | |
| file | InterRepeatContactFilter.cc |
| file | InterRepeatContactFilter.fwd.hh |
| file | InterRepeatContactFilter.hh |
| file | InterRepeatContactFilterCreator.hh |
| file | IntraRepeatContactFilter.cc |
| file | IntraRepeatContactFilter.fwd.hh |
| file | IntraRepeatContactFilter.hh |
| file | IntraRepeatContactFilterCreator.hh |
| file | JScoreEvaluator.cc |
| file | JScoreEvaluator.hh |
| file | JScoreEvaluatorCreator.cc |
| file | JScoreEvaluatorCreator.hh |
| Header for JScoreEvaluatorCreator. | |
| file | JumpEvaluator.cc |
| file | JumpEvaluator.hh |
| file | JumpNrEvaluatorCreator.cc |
| file | JumpNrEvaluatorCreator.hh |
| Header for JumpNrEvaluatorCreator. | |
| file | LeastNativeLike9merFilter.cc |
| file | LeastNativeLike9merFilter.fwd.hh |
| file | LeastNativeLike9merFilter.hh |
| Find the rmsd to the worst 9mer in the chain. | |
| file | LeastNativeLike9merFilterCreator.hh |
| file | LongestContinuousApolarSegmentFilter.cc |
| This filter computes the longest continuous stretch of apolar residues within a pose or selection. | |
| file | LongestContinuousApolarSegmentFilter.fwd.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousApolarSegmentFilter.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousApolarSegmentFilterCreator.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousPolarSegmentFilter.cc |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousPolarSegmentFilter.fwd.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousPolarSegmentFilter.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | LongestContinuousPolarSegmentFilterCreator.hh |
| This filter computes the longest continuous stretch of polar residues within a pose or selection. | |
| file | MembAccesResidueLipophilicityFilter.cc |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | MembAccesResidueLipophilicityFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | MembAccesResidueLipophilicityFilter.hh |
| calcualtes the overall lipophilicity of the pose for each residue exposed to the ocre, the dsTbL membrane insertion energy is computed, and multiplied by the relative SASA of that residue. this amounts, generally speaking, ot the energy of isnertion of the protein, in that conformation. useful for filtering poses that are not hydrophobic enough | |
| file | MembAccesResidueLipophilicityFilterCreator.hh |
| FilterCreators for the MembAccesResidueLipophilicityFilterCreator. | |
| file | MotifScoreFilter.cc |
| Scores poses using will's motifScore. | |
| file | MotifScoreFilter.fwd.hh |
| file | MotifScoreFilter.hh |
| file | MotifScoreFilterCreator.hh |
| file | MPSpanAngleFilter.cc |
| calculates the angle between the TM span and the membrane normal | |
| file | MPSpanAngleFilter.fwd.hh |
| calculates the angle between the TM span and the membrane normal | |
| file | MPSpanAngleFilter.hh |
| definition of filter class MPSpanAngleFilter. | |
| file | MPSpanAngleFilterCreator.hh |
| file | MutationsFilter.cc |
| file | MutationsFilter.fwd.hh |
| forward declaration for MutationsFilter | |
| file | MutationsFilter.hh |
| A filter to return the number of mutated positions within a given set of designable resdues. | |
| file | MutationsFilterCreator.hh |
| FilterCreators for the MutationsFilter. | |
| file | NativeEvaluatorCreator.cc |
| file | NativeEvaluatorCreator.hh |
| Header for NativeEvaluatorCreator. | |
| file | NeighborTypeFilter.cc |
| file | NeighborTypeFilter.fwd.hh |
| file | NeighborTypeFilter.hh |
| definition of filter class InterfaceSasaFilter. | |
| file | NeighborTypeFilterCreator.hh |
| FilterCreators for the NeighborTypeFilterCreator. | |
| file | NetChargeFilter.cc |
| file | NetChargeFilter.fwd.hh |
| file | NetChargeFilter.hh |
| definition of filter class NetChargeFilter. | |
| file | NetChargeFilterCreator.hh |
| FilterCreators for the NetChargeFilterCreator. | |
| file | NMerPSSMEnergyFilter.cc |
| file | NMerPSSMEnergyFilter.fwd.hh |
| file | NMerPSSMEnergyFilter.hh |
| definition of filter class NMerPSSMEnergyFilter. | |
| file | NMerPSSMEnergyFilterCreator.hh |
| FilterCreators for the NMerPSSMEnergyFilter. | |
| file | NMerSVMEnergyFilter.cc |
| file | NMerSVMEnergyFilter.fwd.hh |
| file | NMerSVMEnergyFilter.hh |
| definition of filter class NMerSVMEnergyFilter. | |
| file | NMerSVMEnergyFilterCreator.hh |
| FilterCreators for the NMerSVMEnergyFilter. | |
| file | NonSequentialNeighborsFilter.cc |
| file | NonSequentialNeighborsFilter.fwd.hh |
| file | NonSequentialNeighborsFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | NonSequentialNeighborsFilterCreator.hh |
| FilterCreators for the NonSequentialNeighborsFilter. | |
| file | PackerNeighborGraphFilter.cc |
| file | PackerNeighborGraphFilter.hh |
| header file for packer neighbor graph based filter | |
| file | PackStatFilter.cc |
| file | PackStatFilter.fwd.hh |
| file | PackStatFilter.hh |
| header file for PackStatFilter class. | |
| file | PackStatFilterCreator.hh |
| FilterCreator for the PackStatFilter. | |
| file | PalesEvaluator.cc |
| file | PalesEvaluator.hh |
| file | PalesEvaluatorCreator.cc |
| file | PalesEvaluatorCreator.hh |
| Header for PalesEvaluatorCreator. | |
| file | PDDFScoreFilter.cc |
| runs reject or accept filters on pose | |
| file | PDDFScoreFilter.fwd.hh |
| file | PDDFScoreFilter.hh |
| file | PoolEvaluatorCreator.cc |
| file | PoolEvaluatorCreator.hh |
| Header for PoolEvaluatorCreator. | |
| file | PoseCommentFilter.cc |
| file | PoseCommentFilter.fwd.hh |
| file | PoseCommentFilter.hh |
| file | PoseCommentFilterCreator.hh |
| FilterCreators for the PoseCommentFilter. | |
| file | PoseInfoFilter.cc |
| file | PoseInfoFilter.hh |
| Filter for outputing information about the pose. | |
| file | PoseInfoFilterCreator.hh |
| FilterCreator for the PoseInfoFilter. | |
| file | PoseMetricEvaluator.fwd.hh |
| Forward header for PoseMetricEvaluator. | |
| file | PoseMetricEvaluator.hh |
| file | PredictedBurialEvaluator.cc |
| file | PredictedBurialEvaluator.hh |
| file | PredictedBurialFnEvaluatorCreator.cc |
| file | PredictedBurialFnEvaluatorCreator.hh |
| Header for PredictedBurialFnEvaluatorCreator. | |
| file | RangeFilter.cc |
| file | RangeFilter.fwd.hh |
| file | RangeFilter.hh |
| Range filter evaluates if the return value of a particular filter is between a specific range. | |
| file | RangeFilterCreator.hh |
| FilterCreator for the RangeFilter. | |
| file | RDC_Evaluator.cc |
| file | RDC_Evaluator.fwd.hh |
| file | RDC_Evaluator.hh |
| file | RdcEvaluatorCreator.cc |
| file | RdcEvaluatorCreator.hh |
| Header for RdcEvaluatorCreator. | |
| file | RdcSelectEvaluatorCreator.cc |
| file | RdcSelectEvaluatorCreator.hh |
| Header for RdcSelectEvaluatorCreator. | |
| file | RdcTargetEvaluatorCreator.cc |
| file | RdcTargetEvaluatorCreator.hh |
| Header for RdcTargetEvaluatorCreator. | |
| file | ReadPoseExtraScoreFilter.cc |
| file | ReadPoseExtraScoreFilter.fwd.hh |
| file | ReadPoseExtraScoreFilter.hh |
| definition of filter class ReadPoseExtraScoreFilter. | |
| file | ReadPoseExtraScoreFilterCreator.hh |
| FilterCreators for the ReadPoseExtraScoreFilter. | |
| file | RepeatParameterFilter.cc |
| can filter a pose for the rise, run and omega. Many of the functions have been stolen from RepeatGlobalFrame but made const to work with filter obejcts. | |
| file | RepeatParameterFilter.fwd.hh |
| forward declaration for RepeatParameterFilter | |
| file | RepeatParameterFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | RepeatParameterFilterCreator.hh |
| file | ReportFilter.cc |
| file | ReportFilter.fwd.hh |
| file | ReportFilter.hh |
| Simple filter that tests whether a file exists. Useful to test whether we're recovering from a checkpoint. | |
| file | ReportFilterCreator.hh |
| FilterCreators for the ReportFilter. | |
| file | ResidueBurialFilter.cc |
| file | ResidueBurialFilter.fwd.hh |
| file | ResidueBurialFilter.hh |
| definition of filter class ResidueBurialFilter. | |
| file | ResidueBurialFilterCreator.hh |
| FilterCreator for the ResidueBurialFilter. | |
| file | ResidueChiralityFilter.cc |
| checks the chirality of a specific residues, whether it is D or L | |
| file | ResidueChiralityFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | ResidueChiralityFilter.hh |
| checks the chirality of a specific residues, whether it is D or L | |
| file | ResidueChiralityFilterCreator.hh |
| FilterCreators for the ResidueChiralityFilterCreator. | |
| file | ResidueCountFilter.cc |
| Filter on the total number of residues in the structure. | |
| file | ResidueCountFilter.fwd.hh |
| Filter on the total number of residues in the structure. | |
| file | ResidueCountFilter.hh |
| Filter on the total number of residues in the structure. | |
| file | ResidueCountFilterCreator.hh |
| FilterCreators for the ResidueCountFilter. | |
| file | ResidueDepthFilter.cc |
| file | ResidueDepthFilter.fwd.hh |
| file | ResidueDepthFilter.hh |
| file | ResidueDepthFilterCreator.hh |
| FilterCreator for the ResidueDepthFilter. | |
| file | ResidueDistanceFilter.cc |
| file | ResidueDistanceFilter.fwd.hh |
| file | ResidueDistanceFilter.hh |
| definition of filter class ResidueDistanceFilter. | |
| file | ResidueDistanceFilterCreator.hh |
| FilterCreator for the ResidueDistanceFilter. | |
| file | ResidueIEFilter.cc |
| file | ResidueIEFilter.fwd.hh |
| file | ResidueIEFilter.hh |
| definition of filter class ResidueIEFilter. | |
| file | ResidueIEFilterCreator.hh |
| FilterCreators for the ResidueIEFilterCreator. | |
| file | ResidueLipophilicityFilter.cc |
| returns the value calculated by the energy term mp_res_lipo. useful for understanding and debugging this energy term. use the output_file tag to print a full table of parameters from within the energy term | |
| file | ResidueLipophilicityFilter.fwd.hh |
| returns the value calculated by the energy term mp_res_lipo. useful for understanding and debugging this energy term. use the output_file tag to print a full table of parameters from within the energy term | |
| file | ResidueLipophilicityFilter.hh |
| definition of filter class ResidueLipophilicityFilter. | |
| file | ResidueLipophilicityFilterCreator.hh |
| FilterCreators for the ResidueLipophilicityFilterCreator. | |
| file | ResidueSelectionDistanceFilter.cc |
| file | ResidueSelectionDistanceFilter.fwd.hh |
| file | ResidueSelectionDistanceFilter.hh |
| loops over the residues in a residue selector and reports the average distance | |
| file | ResidueSelectionDistanceFilterCreator.hh |
| FilterCreator for the ResidueSelectionDistanceFilter. | |
| file | ResidueSetChainEnergyFilter.cc |
| file | ResidueSetChainEnergyFilter.fwd.hh |
| file | ResidueSetChainEnergyFilter.hh |
| definition of filter class ResidueSetChainEnergyFilter. | |
| file | ResidueSetChainEnergyFilterCreator.hh |
| FilterCreators for the ResidueSetChainEnergyFilter. | |
| file | ResiduesInInterfaceFilter.cc |
| file | ResiduesInInterfaceFilter.fwd.hh |
| file | ResiduesInInterfaceFilter.hh |
| Reports to Tracer which residues are designable in a taskfactory. | |
| file | ResiduesInInterfaceFilterCreator.hh |
| FilterCreators for the ResiduesInInterfaceFilter. | |
| file | RGFilter.cc |
| runs reject or accept filters on pose | |
| file | RGFilter.hh |
| file | RmsdEvaluator.cc |
| file | RmsdEvaluator.hh |
| file | RmsdEvaluatorCreator.cc |
| file | RmsdEvaluatorCreator.hh |
| Header for RmsdEvaluatorCreator. | |
| file | RmsdTargetEvaluatorCreator.cc |
| file | RmsdTargetEvaluatorCreator.hh |
| Header for RmsdTargetEvaluatorCreator. | |
| file | SaveResfileToDiskFilter.cc |
| Outputs a resfile based on a given set of taskoperations. | |
| file | SaveResfileToDiskFilter.fwd.hh |
| file | SaveResfileToDiskFilter.hh |
| header file for SaveResfileToDiskFilter class | |
| file | SaveResfileToDiskFilterCreator.hh |
| FilterCreator for the SaveResfileToDiskFilter. | |
| file | SAXSScoreFilter.cc |
| runs reject or accept filters on pose | |
| file | SAXSScoreFilter.fwd.hh |
| file | SAXSScoreFilter.hh |
| file | ScoreEvaluator.cc |
| file | ScoreEvaluator.hh |
| file | SecretionPredictionFilter.cc |
| file | SecretionPredictionFilter.fwd.hh |
| Filter for greasy helices https://www.nature.com/articles/nature06387. | |
| file | SecretionPredictionFilter.hh |
| Filter for greasy helices https://www.nature.com/articles/nature06387. | |
| file | SecretionPredictionFilterCreator.hh |
| file | SequenceDistanceFilter.cc |
| file | SequenceDistanceFilter.fwd.hh |
| file | SequenceDistanceFilter.hh |
| file | SequenceDistanceFilterCreator.hh |
| FilterCreators for the SequenceDistanceFilter. | |
| file | ShapeComplementarityFilter.cc |
| file | ShapeComplementarityFilter.fwd.hh |
| file | ShapeComplementarityFilter.hh |
| header file for ShapeComplementarityFilter class | |
| file | ShapeComplementarityFilterCreator.hh |
| FilterCreator for the ShapeComplementarityFilter. | |
| file | SheetFilter.cc |
| runs reject or accept filters on pose | |
| file | SheetFilter.hh |
| file | SidechainRmsdFilter.cc |
| A filter based on automorphic sidechain RMSD. | |
| file | SidechainRmsdFilter.fwd.hh |
| A filter based on automorphic sidechain RMSD. | |
| file | SidechainRmsdFilter.hh |
| A filter based on automorphic sidechain RMSD. | |
| file | SidechainRmsdFilterCreator.hh |
| file | SimpleHbondsToAtomFilter.cc |
| Simple filter for detercting Hbonds to atom with energy < energy cutoff. | |
| file | SimpleHbondsToAtomFilter.fwd.hh |
| Simple filter for detercting Hbonds to atom with energy < energy cutoff. | |
| file | SimpleHbondsToAtomFilter.hh |
| Simple filter for detercting Hbonds to atom with energy < energy cutoff. | |
| file | SimpleHbondsToAtomFilterCreator.hh |
| Simple filter for detercting Hbonds to atom with energy < energy cutoff. | |
| file | SimpleMetricFilter.cc |
| A filter takes any RealMetric and applies a set cutoff to filter the model. | |
| file | SimpleMetricFilter.fwd.hh |
| A filter takes any RealMetric and applies a set cutoff to filter the model. | |
| file | SimpleMetricFilter.hh |
| A filter takes any RealMetric and applies a set cutoff to filter the model. | |
| file | SimpleMetricFilterCreator.hh |
| A filter takes any RealMetric and applies a set cutoff to filter the model. | |
| file | SpanTopologyMatchPoseFilter.cc |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | SpanTopologyMatchPoseFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | SpanTopologyMatchPoseFilter.hh |
| definition of filter class SpanTopologyMatchPoseFilter. | |
| file | SpanTopologyMatchPoseFilterCreator.hh |
| FilterCreators for the SpanTopologyMatchPoseFilterCreator. | |
| file | SSElementLengthFilter.cc |
| file | SSElementLengthFilter.fwd.hh |
| filter structures by longest,shortest or avg length of a given secondary structure type | |
| file | SSElementLengthFilter.hh |
| filter structures by longest,shortest or avg length of a given secondary structure type | |
| file | SSElementLengthFilterCreator.hh |
| file | SSElementMotifContactFilter.cc |
| file | SSElementMotifContactFilter.fwd.hh |
| Filter for the interconnection between secondary elements. | |
| file | SSElementMotifContactFilter.hh |
| Reports either the average ss-contact or the worst ss-contacting element. | |
| file | SSElementMotifContactFilterCreator.hh |
| file | SSMotifFinderFilter.cc |
| file | SSMotifFinderFilter.fwd.hh |
| file | SSMotifFinderFilter.hh |
| file | SSMotifFinderFilterCreator.hh |
| FilterCreators for the SSMotifFinderFilter. | |
| file | StemFinderFilter.cc |
| file | StemFinderFilter.fwd.hh |
| file | StemFinderFilter.hh |
| file | StemFinderFilterCreator.hh |
| FilterCreators for the StemFinderFilter. | |
| file | StructuralSimilarityEvaluator.cc |
| file | StructuralSimilarityEvaluator.fwd.hh |
| forward declaration for StructuralSimilarityEvaluator class. | |
| file | StructuralSimilarityEvaluator.hh |
| file | StructureSimilarityEvaluatorCreator.cc |
| file | StructureSimilarityEvaluatorCreator.hh |
| Header for StructureSimilarityEvaluatorCreator. | |
| file | SymmetricMotifFilter.cc |
| file | SymmetricMotifFilter.hh |
| file | SymmetricMotifFilterCreator.hh |
| file | TaskAwareSASAFilter.cc |
| Calculates SASA for a set of residues defined by TaskOperations. | |
| file | TaskAwareSASAFilter.fwd.hh |
| file | TaskAwareSASAFilter.hh |
| header file for TaskAwareSASAFilter class | |
| file | TaskAwareSASAFilterCreator.hh |
| FilterCreator for the TaskAwareSASAFilter. | |
| file | TaskAwareScoreTypeFilter.cc |
| file | TaskAwareScoreTypeFilter.fwd.hh |
| forward declaration for TaskAwareScoreTypeFilter | |
| file | TaskAwareScoreTypeFilter.hh |
| file | TaskAwareScoreTypeFilterCreator.hh |
| FilterCreators for the TaskAwareScoreTypeFilter. | |
| file | TerminusDistanceFilter.cc |
| file | TerminusDistanceFilter.fwd.hh |
| file | TerminusDistanceFilter.hh |
| definition of filter class TerminusDistanceFilter. | |
| file | TerminusDistanceFilterCreator.hh |
| FilterCreators for the TerminusDistanceFilterCreator. | |
| file | TMsAACompFilter.cc |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | TMsAACompFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | TMsAACompFilter.hh |
| definition of filter class TMsAACompFilter. | |
| file | TMsAACompFilterCreator.hh |
| FilterCreators for the TMsAACompFilterCreator. | |
| file | TMsSpanMembraneFilter.cc |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | TMsSpanMembraneFilter.fwd.hh |
| return the matching (%) between span file determined topology and of the actual pose | |
| file | TMsSpanMembraneFilter.hh |
| definition of filter class TMsSpanMembraneFilter. | |
| file | TMsSpanMembraneFilterCreator.hh |
| FilterCreators for the TMsSpanMembraneFilterCreator. | |
| file | TotalSasaFilter.cc |
| file | TotalSasaFilter.fwd.hh |
| file | TotalSasaFilter.hh |
| definition of filter class TotalSasaFilter. | |
| file | TotalSasaFilterCreator.hh |
| FilterCreators for the TotalSasaFilterCreator. | |
 1.8.7
1.8.7