Documentation for the PyRosetta Toolkit GUI
Author:
Jared Adolf-Bryfogle (jadolfbr@gmail.com)
!
Metadata
Deprecated
Last edited 10/1/18. Code by Jared Adolf-Bryfogle < jadolfbr@gmail.com >.
The PI for this application is Roland Dunbrack < Roland.Dunbrack@fccc.edu >
Code and Demo
The PyRosetta Toolkit has not been ported to PyRosetta-4 unfortunately and is only distributed and comparable with PyRosetta-3.
The application is available in PyRosetta-3 in app/pyrosetta_toolkit
PyRosetta Setup:
Optional PyMOL Integration Setup:
Optional SCWRL Integration Setup:
- Obtain SCWRL from: http://dunbrack.fccc.edu/scwrl4/
- Copy the exe/binary into the
pyrosetta/apps/pyrosetta_toolkit/SCWRL directory in the respective OS directory.
Run the program:
- Use
pyrosetta_toolkitafter running SetPyRosettaEnvironment.sh (creates an alias).
References
Jared Adolf-Bryfogle and Roland Dunbrack (2013) "The PyRosetta Toolkit: A Graphical User Interface for the Rosetta Software Suite. " PloS-ONE RosettaCON2012 special collection
Purpose
This GUI is used to setup Rosetta specific filetypes, do interactive modeling using Rosetta as its base, run rosetta protocols, and analyze decoy results.
Limitations
This application does not run all Rosetta Applications. It does, however, run the most common ones. Although multiprocessing is implemented, full production runs of many rosetta applications usually require a cluster. Always check relevant Rosetta Documentation on RosettaCommons. Options for all protocols can be added through the Option System Manager or through it's protocol-specific UI. Symmetry is not supported at this time.
Menus
File Menu
Load a pose through this menu first either from a file or directly from the RCSB. If you have PyMOL setup, it will send the pose to PyMOL after it loads into Rosetta. Loading a pose will open up the PDB Cleaning window by default to prep the PDB for import into Rosetta.
PDB Cleaning Functions:
- renaming waters
- removing waters
- removing hetatm
- renaming DNA residues
- renaming MD atoms
- changing occupancies to 1.0
- renumbering pose from 1 + accounting for insertions
checking that all residues are able to be read by Rosetta
Current Imports:GUI Session, Param PathList, Loop File
- Current Exports:GUI Session, Param PathList, Loop File, basic Resfile, basic Blueprint, SCWRL seq file, FASTA (pose + regions)
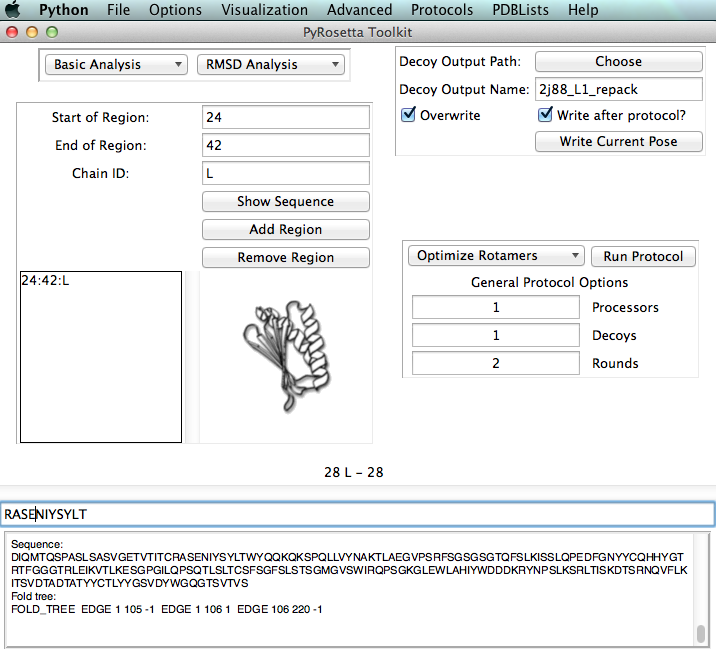
Main Window
The main window is used to run common protocols, some anlysis, setup decoy output, and finally set a region.
The default region is the whole protein. To specify loops for loop modeling, regions for minimization or residues for repacking, etc. add a region:
- Chain: can be added by just specifying the chain
- N-terminus:Leave 'start' entry box blank, or enter any string, specify both chain and end residue number
- C-terminus:Leave 'end' entry box blank, or enter any string, specify both chain and start residue number
- Residue: Have start + end be the same residue.
Sequence:
- A sequence of the pose or region can be found in the lower part of the screen. Placing your cursor at the left part of a residue will show its residue number, chain, and rosetta resnum. This can be colored in pymol and used to select residues in the Per-Residue Control Window described below.
Output Options:
- The default output directory is the dir of the current pose. This and other output options for decoys can be set in the upper right and lower right areas of the main window.
- Multiple Processors are used only for protocol runs of >1 decoy, as well as some functions from the PDBList menu.
Protocols:
- A few common minimization protocols can be ran through the main window in addition to the protocols menu (Repack, Minimize, Relax, SCWRL).
Options Menu
- Option System manager:
- : : Setup Rosetta command-line options using the options system manager
- Configure Score Function:
- : : Default Scorefunction is score12prime. The scorefunction set here is used by all protocols. See the protocols section for more information. Please use this window to change the scorefunction or the weights of any terms.
- : : More weight sets can be given in this window by selecting options->show ALL scorefunctions
- Set General Protocol Options:
- : : A number of rounds can be set for all protocols. By default, if more than one round is set, the boltzmann criterion is applied to the pose after each round using the current scorefunction. This is especially useful for packing or design. Here, you can control the temperature, and the behavior of what happens each round and if the lowest energy pose is recovered at the end of the run.
Visualization Menu
!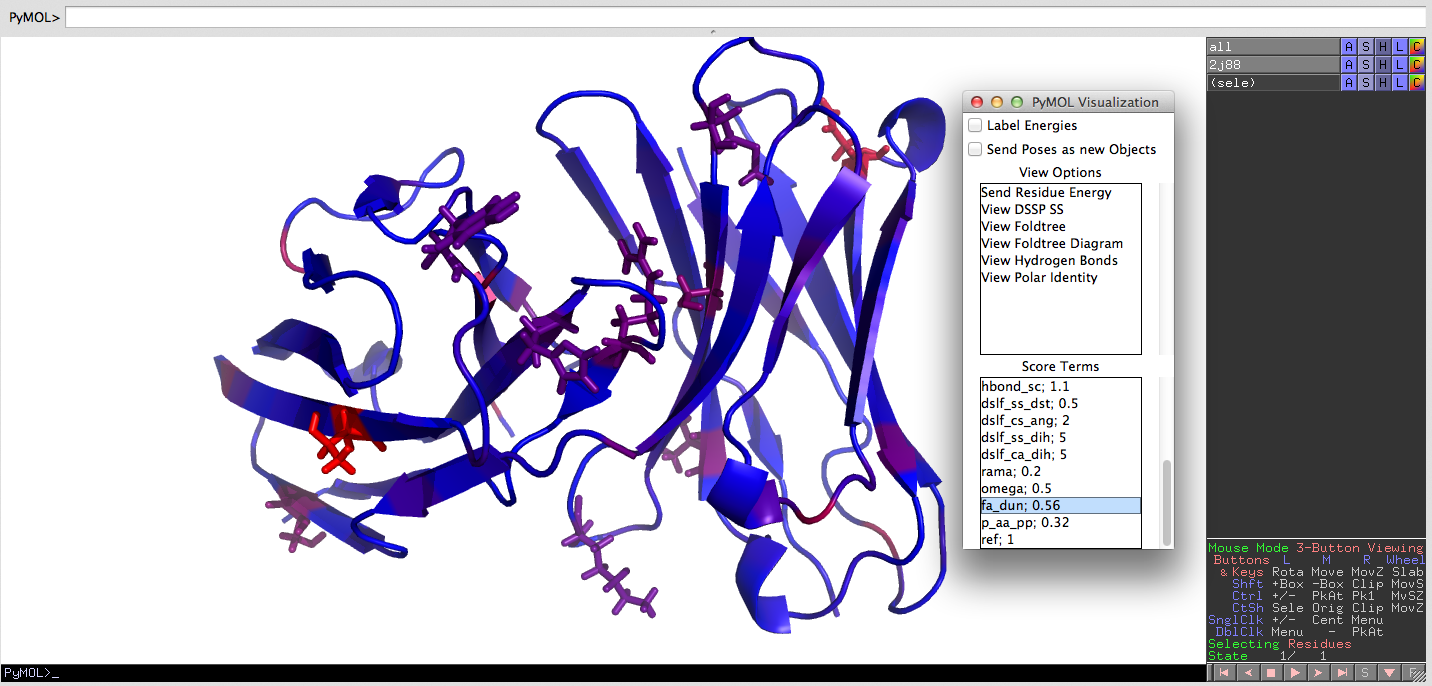 PyMOL Integration through the PyMOL Mover. A pose can be shown in PyMOL at any time. Regions can be colored, and the PyMOL Observer can be enabled that updates pymol upon structural and energetic changes to the current pose.
PyMOL Integration through the PyMOL Mover. A pose can be shown in PyMOL at any time. Regions can be colored, and the PyMOL Observer can be enabled that updates pymol upon structural and energetic changes to the current pose.
- Advanced PyMOL Visualization button
- : : This is a simple window that can be used while using PyMOL. It is small so that you can incorporate it into your modeling workflow.
- : : For energies, blue is low energy and red is high energy. Spectrum cannot be changed at this time, but will be added in the future.
- : : Labels for energies can be useful, although slow. Use the PyMOL commands to remove them once sent.
Advanced Menu
The Ligand/NCAA/PTM Manager
- Many non-cannonicals are off-by-default for Rosetta, and this window will allow you to enable them and also design residues into them.
- Energy function optimization is also given here. Since the default electrostatic score term in the default scorefunction is statistical in nature, it will not work for non-cannoicals. At the very least, changing this to the coulumbic score term is a more physically realistic representation of the sytem.
- The mm_std scorefunction ( Douglas Renfrew, Eun Jung Choi, Brian Kuhlman, 'Using Noncanonical Amino Acids in Computational Protein-Peptide Interface Design' (2011) PLoS One. ) is currently undergoing broad tests for proteins and other systems, but seems to work very will for NCAA design and rotamers.
- The orbitals scorefunction, developed by Steven Combs et al (Meiler Lab) is also a great alternative, which is residue independant. Tests are ongoing for all of these scorefunction changes.
Design Toolbox
- This is a window which will allow you to build a resfile for the loaded pose. You can do individual residues or a region of the pose. See the help menu or paper for information on what the values are in this window.
- The left most box is for residue categories. This is for easy selection and design. Use the etc category for typical rosetta resfile options (NATRO,NATAA,ALLAA). Clicking one of the categories will populate the right box. Here you actually choose the residues types you want to open by double clicking them. Selecting these will limit the design to only those chosen. Residues you have chosen then go into the lower most box. Changing the residue number you have selected, will change what goes in this box so you can edit your current design. Double clicking a residue in the box will remove it.
- Click the Write Resfile button to open a save as dialog.
Enable constraints
- Will prompt for a constraint file. For more information, please see the rosettacommons manual.
Insert data into B factor Column
- This is mainly an analysis utility, but it can be useful for data visualization, especially for publications.
- Data should be in a text or csv file, deliminated by either a space, tab, column, comma.
- Data should have at least 3 columns: pdb resnum, chain, data to insert, optional atom name
- The UI for this utility function will allow you choose the colunns for each required piece as well as the deliminator. It will use your currently loaded pose and output a new pose with the data in the B-factor column. Load the pose into PyMOL or your favorite visualizer to see the data.
Per Residue control and Analysis
- This is a great interactive modeling window. Here you can do many modeling tasks including mutating individual residues, add variants such as phosphorylations/acetylations, repack rotamers, chaing dihedrals, etc. You can also analze rotamer probabilies and individual residue energies.
Analysis Movers:
- Interface Analyzer
- Loop Analyzer
- Packing Analyzer
- VIP
Protocols Menu
- A loaded pose is required for all protocols. Options not given in the main UI or their protocol-specific UI can be set using the options system manager. Please refer to RosettaCommons manual for all protocols, as well as the references listed there and within the GUI.
- The default scorefunction is score12prime. Ligands/NCAA/PTM may require modifications to the scorefunction. Please use the advanced menu to make these modifications. Centroid-based protocols and other specific protocols require different scorefunctions. They are given below, and you will be prompted in the GUI if these are not set.
Currently Implemented:
High/Low Resolution Docking
- Will prompt for docking partners using the same notation as the docking application.
- ScoreFunction: Use interchain_cen scorefunction for low-resolution and docking for high-resolution.
High/Low Resolution Loop Modeling (KIC/CCD)
- Will prompt for fragset(CCD)/ extend options.
- Default cutpoint is around the middle of the loop.
- ScoreFunction: Use centroid + score4L patch for low-resolution.
Fixedbb Design (UI)
- Will prompt for a resfile.
Floppytail (UI)
Grafting (UI)
- Graft a region from one protein into another.
- Many options are given. All are described in the UI. Choosing a graft method will give a description of that method.
- It is assumed that the loaded pose is the one you will graft into. You can pick the doner pose from the UI. They can both be the same PDB file.
- If the region is not already deleted, it will delete it upon grafting.
FastRelax
ClassicRelax
PackRotamers (Rosetta)
PackRotamers (SCWRL)
Minimizer
- The Rosetta default minimizer is dfpmin. Use the options system to set a different one setting '-run:min_tolerance'
- The Relax application uses dfpmin_armijo_nonmonotone. Visit Minimization Overview for more information.
- The Rosetta default tolerance is 0.000001. Use the options system to set a different value by setting -run:min_tolerance.
Server Links:
- ROSIE
- Fragments
- Backrub
- Interface Alanine Scan
- DNA Interface Scan
- Scaffold Select
PDBList Menu
A PDBList is simply a list of full paths to PDB files, one on each line. It is used by the GUI to analyze decoys. Methods for making and using a PDBList reside here.
Main Functions:
- Lowest Energy - get lowest energy poses from a PDBList
- Energy vs RMSD - Calculate Energy vs RMSD + output a file that can be graphed in any graphing application.
- Output FASTA - Output FASTA with sequence for each pose or only of the given region
- Design Statistics - Use FASTA to calculate design stats from a Rosetta design run. Outputs textfiles and an R plot. Needs reference pose loaded. Does not work with insertions and deletions during the design yet.
Tips
- -Set default (command-line) Options through the options system manager window
- -Set default Scorefunction through the scorefunction window - Used by all protocols.
- -Save and Load GUI Sessions using the file menu
- -Advanced Users: To add personal windows and functions to the GUI, see developer html in pyrosetta_toolkit/documentation directory.
Bashrc Setup
This is so you do not need to source SetPyRosettaEnvironment.sh each time you want to use Toolkit. Takes a few minutes, but worth it. For more information on to what a bash shell is: http://superuser.com/questions/49289/what-is-bashrc-file
To do this:
- Add a line to your bashrc or zshrc if your using zsh (\$HOME/.bashrc) or (\$HOME/.bash_profile on mac) (\$HOME/.zshrc if you use zsh shell):
source full/path/to/SetPyRosettaEnvironment.sh(This file is found in the PyRosetta directory)
PyMOL Setup
Setup so that the pymol server runs + listens each time PyMOL is opened.
- Add a line in the file \$HOME/.pymolrc (may be a new file):
run full/path/to/PyMOLPyRosettaServer.py(This file is found in the PyRosetta directory)
Changes since last release
This will be the first release