Directory dependency graph for energy_methods:
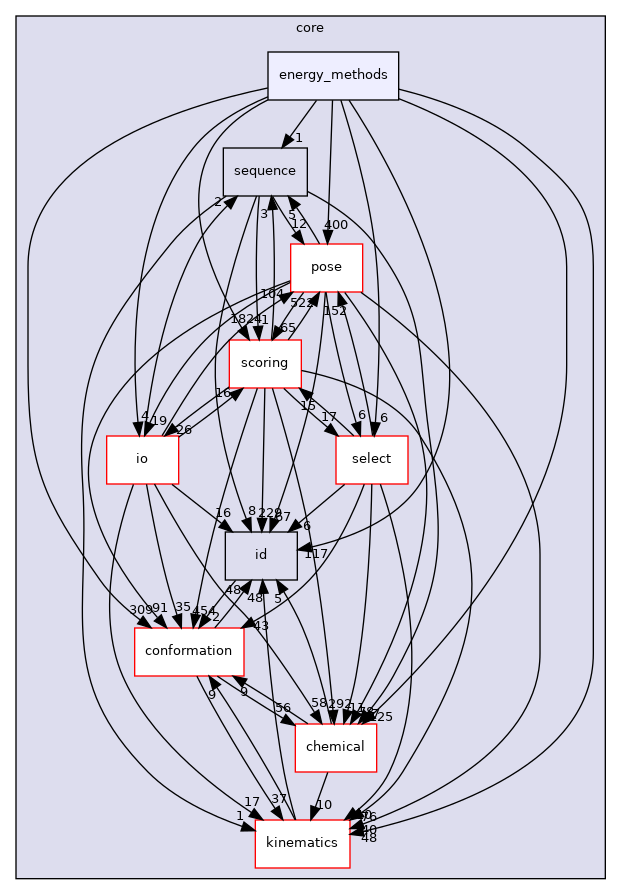
Files | |
| file | AACompositionEnergy.cc |
| file | AACompositionEnergy.fwd.hh |
| file | AACompositionEnergy.hh |
| file | AACompositionEnergyCreator.hh |
| file | AARepeatEnergy.cc |
| file | AARepeatEnergy.fwd.hh |
| file | AARepeatEnergy.hh |
| file | AARepeatEnergyCreator.hh |
| file | AbegoEnergy.cc |
| ABEGO energy method class implementation. | |
| file | AbegoEnergy.fwd.hh |
| ABEGO energy method class forward declaration. | |
| file | AbegoEnergy.hh |
| file | AbegoEnergyCreator.hh |
| file | ArgCationPiEnergy.cc |
| Cation pi term that specializes in bringing Arginine and rings together. | |
| file | ArgCationPiEnergy.fwd.hh |
| Cation pi term that specializes in bringing Arginine and rings together. | |
| file | ArgCationPiEnergy.hh |
| Cation pi term that specializes in bringing Arginine and rings together. | |
| file | ArgCationPiEnergyCreator.hh |
| file | AromaticBackboneRestraintEnergy.cc |
| Aramid backbone restraint energy method class implementation. | |
| file | AromaticBackboneRestraintEnergy.fwd.hh |
| Noncanonical ring closure energy method class forward declaration. | |
| file | AromaticBackboneRestraintEnergy.hh |
| Aromatic backbone restraint energy method class headers. | |
| file | AromaticBackboneRestraintEnergyCreator.hh |
| Declaration for the class that connects AromaticBackboneRestraintEnergy with the ScoringManager. | |
| file | AspartimidePenaltyEnergy.cc |
| file | AspartimidePenaltyEnergy.fwd.hh |
| Forward declarations for the AspartimidePenaltyEnergy. | |
| file | AspartimidePenaltyEnergy.hh |
| This is a score term that penalizes sequences that are likely to result in aspartimide formation during peptide synthesis. | |
| file | AspartimidePenaltyEnergyCreator.hh |
| Declaration for the class that connects AspartimidePenaltyEnergy with the ScoringManager. | |
| file | BranchEnergy.cc |
| Method definitions for BranchEnergyCreator and BranchEnergy classes. | |
| file | BranchEnergy.fwd.hh |
| file | BranchEnergy.hh |
| Method declarations and typedefs for BranchEnergy. | |
| file | BranchEnergyCreator.hh |
| Declaration for the class that connects BranchEnergy with the ScoringManager. | |
| file | Burial_v2Energy.cc |
| file | Burial_v2Energy.hh |
| energy term use for score burial of a specific residue | |
| file | Burial_v2EnergyCreator.hh |
| Declaration for the class that connects Burial_v2Energy with the ScoringManager. | |
| file | BurialEnergy.cc |
| file | BurialEnergy.hh |
| energy term use for scoring predicted burial | |
| file | BurialEnergyCreator.hh |
| Declaration for the class that connects BurialEnergy with the ScoringManager. | |
| file | CartesianBondedEnergy.cc |
| Harmonic bondangle/bondlength/torsion constraints. | |
| file | CartesianBondedEnergy.fwd.hh |
| file | CartesianBondedEnergy.hh |
| file | CartesianBondedEnergyCreator.hh |
| Declaration for the class that connects CartesianBondedEnergy with the ScoringManager. | |
| file | CCS_IMMSEnergy.cc |
| file | CCS_IMMSEnergy.fwd.hh |
| file | CCS_IMMSEnergy.hh |
| Forward model based off of CCS data from Ion Mobility Mass Spec Data. | |
| file | CCS_IMMSEnergyCreator.hh |
| Declaration for the class that connects CCS_IMMSEnergy with the ScoringManager. | |
| file | CenHBEnergy.cc |
| Smooth, differentiable version of centroid hbond term. | |
| file | CenHBEnergy.fwd.hh |
| file | CenHBEnergy.hh |
| Smooth, differentiable version of centroid hbond term. | |
| file | CenHBEnergyCreator.hh |
| Declaration for the class that connects CenHBEnergy with the ScoringManager. | |
| file | CenPairEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | CenPairEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | CenPairEnergyCreator.hh |
| file | CenPairMotifDegreeEnergy.cc |
| file | CenPairMotifDegreeEnergy.hh |
| file | CenPairMotifDegreeEnergyCreator.hh |
| file | CenPairMotifEnergy.cc |
| file | CenPairMotifEnergy.hh |
| file | CenPairMotifEnergyCreator.hh |
| file | CenRotEnvEnergy.cc |
| CenRot version of centroid env. | |
| file | CenRotEnvEnergy.hh |
| file | CenRotEnvEnergyCreator.hh |
| file | CenRotPairEnergy.cc |
| CenRot version of cen pair. | |
| file | CenRotPairEnergy.hh |
| file | CenRotPairEnergyCreator.hh |
| file | CentroidDisulfideEnergy.cc |
| Centroid Disulfide Energy class implementation. | |
| file | CentroidDisulfideEnergy.fwd.hh |
| Centroid Disulfide Energy class forward declaration. | |
| file | CentroidDisulfideEnergy.hh |
| Centroid Disulfide Energy class declaration. | |
| file | CentroidDisulfideEnergyCreator.hh |
| Declaration for the class that connects CentroidDisulfideEnergy with the ScoringManager. | |
| file | ChainbreakEnergy.cc |
| Method definitions for ChainbreakEnergyCreator and ChainbreakEnergy classes. | |
| file | ChainbreakEnergy.fwd.hh |
| file | ChainbreakEnergy.hh |
| Method declarations and typedefs for ChainbreakEnergy. | |
| file | ChainbreakEnergyCreator.hh |
| Declaration for the class that connects ChainbreakEnergy with the ScoringManager. | |
| file | ChemicalShiftAnisotropyEnergy.cc |
| CSA energy - Orientation dependent chemical shift. | |
| file | ChemicalShiftAnisotropyEnergy.fwd.hh |
| file | ChemicalShiftAnisotropyEnergy.hh |
| CSP energy. | |
| file | ChemicalShiftAnisotropyEnergyCreator.hh |
| Declaration for the class that connects ChemicalShiftAnisotropyEnergy with the ScoringManager. | |
| file | ContactOrderEnergy.cc |
| calculates the contact order of a given conformation average sequence. | |
| file | ContactOrderEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | ContactOrderEnergyCreator.hh |
| Declaration for the class that connects ContactOrderEnergy with the ScoringManager. | |
| file | ContextDependentGeometricSolEnergy.cc |
| file | ContextDependentGeometricSolEnergy.fwd.hh |
| file | ContextDependentGeometricSolEnergy.hh |
| file | ContextDependentGeometricSolEnergyCreator.hh |
| Declaration for the class that connects GeometricSolEnergy with the ScoringManager. | |
| file | ContextIndependentGeometricSolEnergy.cc |
| file | ContextIndependentGeometricSolEnergy.fwd.hh |
| file | ContextIndependentGeometricSolEnergy.hh |
| file | ContextIndependentGeometricSolEnergyCreator.hh |
| file | CovalentLabelingEnergy.cc |
| file | CovalentLabelingEnergy.fwd.hh |
| file | CovalentLabelingEnergy.hh |
| energy term use for scoring predicted CovalentLabeling | |
| file | CovalentLabelingEnergyCreator.hh |
| Declaration for the class that connects CovalentLabelingEnergy with the ScoringManager. | |
| file | CovalentLabelingFAEnergy.cc |
| file | CovalentLabelingFAEnergy.fwd.hh |
| file | CovalentLabelingFAEnergy.hh |
| energy term use for scoring predicted CovalentLabelingFA | |
| file | CovalentLabelingFAEnergyCreator.hh |
| Declaration for the class that connects CovalentLabelingFAEnergy with the ScoringManager. | |
| file | CustomAtomPairEnergy.cc |
| Simple EnergyMethod for calculating residue-residue constraints under some interaction cutoff. | |
| file | CustomAtomPairEnergy.fwd.hh |
| Statistically derived rotamer pair potential class forward declaration. | |
| file | CustomAtomPairEnergy.hh |
| file | CustomAtomPairEnergyCreator.hh |
| Declaration for the class that connects CustomAtomPairEnergy with the ScoringManager. | |
| file | D2H_SA_Energy.cc |
| sa energy function definition. | |
| file | D2H_SA_Energy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | D2H_SA_EnergyCreator.hh |
| Declaration for the class that connects D2H_SA_Energy with the ScoringManager. | |
| file | DEEREnergy.cc |
| Score term for data obtained with double electron-electron resonance (DEER) | |
| file | DEEREnergy.fwd.hh |
| file | DEEREnergy.hh |
| Score term for data obtained with double electron-electron resonance (DEER) | |
| file | DEEREnergyCreator.hh |
| Parent class for DEEREnergy structure. | |
| file | DEPC_MS_Energy.cc |
| Score term that uses DEPC MS labeling data for Ser, Thr, and Tyr residues to reward based on SASA value, labeling status, and number of hydrophobic neighboring residues and for labeled His and Lys residues to reward based on SASA value. Corroborates with 2021 manuscript by Biehn, Limpikirati, Vachet, and Lindert. | |
| file | DEPC_MS_Energy.fwd.hh |
| Score term using mass spectrometry DEPC data. | |
| file | DEPC_MS_Energy.hh |
| energy term use for scoring predicted DEPC_MS_Energy | |
| file | DEPC_MS_EnergyCreator.hh |
| Declaration for the class that connects DEPC_MS_Energy with the ScoringManager. | |
| file | DFIRE_Energy.cc |
| file | DFIRE_Energy.fwd.hh |
| file | DFIRE_Energy.hh |
| file | DFIRE_EnergyCreator.hh |
| file | DipolarCouplingEnergy.cc |
| DC energy - Orientation dependent chemical shift. | |
| file | DipolarCouplingEnergy.fwd.hh |
| file | DipolarCouplingEnergy.hh |
| CSP energy. | |
| file | DipolarCouplingEnergyCreator.hh |
| Declaration for the class that connects DipolarCouplingEnergy with the ScoringManager. | |
| file | DirectReadoutEnergy.cc |
| Statistically derived DNA contact potential class implementation. | |
| file | DirectReadoutEnergy.fwd.hh |
| file | DirectReadoutEnergy.hh |
| Statistically derived DNA contact potential class declaration. | |
| file | DirectReadoutEnergyCreator.hh |
| Declaration for the class that connects DirectReadoutEnergy with the ScoringManager. | |
| file | DistanceChainbreakEnergy.cc |
| file | DistanceChainbreakEnergy.fwd.hh |
| file | DistanceChainbreakEnergy.hh |
| file | DistanceChainbreakEnergyCreator.hh |
| Declaration for the class that connects DistanceChainbreakEnergy with the ScoringManager. | |
| file | DisulfideMatchingEnergy.cc |
| Centroid Disulfide Energy class implementation. | |
| file | DisulfideMatchingEnergy.fwd.hh |
| Centroid Disulfide Energy class forward declaration. | |
| file | DisulfideMatchingEnergy.hh |
| Centroid Disulfide Energy class declaration. | |
| file | DisulfideMatchingEnergyCreator.hh |
| Declaration for the class that connects DisulfideMatchingEnergy with the ScoringManager. | |
| file | DNA_BaseEnergy.cc |
| file | DNA_BaseEnergy.fwd.hh |
| file | DNA_BaseEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | DNA_BaseEnergyCreator.hh |
| Declaration for the class that connects DNA_BaseEnergy with the ScoringManager. | |
| file | DNA_DihedralEnergy.cc |
| dna scoring | |
| file | DNA_DihedralEnergy.fwd.hh |
| dna scoring | |
| file | DNA_DihedralEnergy.hh |
| dna scoring | |
| file | DNA_DihedralEnergyCreator.hh |
| dna scoring | |
| file | DNA_EnvPairEnergy.cc |
| dna scoring | |
| file | DNA_EnvPairEnergy.fwd.hh |
| dna scoring | |
| file | DNA_EnvPairEnergy.hh |
| dna scoring | |
| file | DNA_EnvPairEnergyCreator.hh |
| dna scoring | |
| file | DNA_ReferenceEnergy.cc |
| dna scoring | |
| file | DNA_ReferenceEnergy.fwd.hh |
| dna scoring | |
| file | DNA_ReferenceEnergy.hh |
| dna scoring | |
| file | DNA_ReferenceEnergyCreator.hh |
| dna scoring | |
| file | DNAChiEnergy.cc |
| file | DNAChiEnergy.fwd.hh |
| file | DNAChiEnergy.hh |
| file | DNAChiEnergyCreator.hh |
| Declaration for the class that connects DNAChiEnergy with the ScoringManager. | |
| file | DNATorsionEnergy.cc |
| DNATorsion energy method class implementation. | |
| file | DNATorsionEnergy.fwd.hh |
| file | DNATorsionEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | DNATorsionEnergyCreator.hh |
| Declaration for the class that connects DNATorsionEnergy with the ScoringManager. | |
| file | DumpTrajectoryEnergy.cc |
| An EnergyMethod that dumps trajectories to file. | |
| file | DumpTrajectoryEnergy.fwd.hh |
| Forward declarations for an EnergyMethod that dumps trajectories to file. | |
| file | DumpTrajectoryEnergy.hh |
| Headers for an EnergyMethod that dumps trajectories to file. | |
| file | DumpTrajectoryEnergyCreator.hh |
| Creator for an EnergyMethod that dump trajectories to file. | |
| file | ElecDensAllAtomCenEnergy.cc |
| file | ElecDensAllAtomCenEnergy.hh |
| file | ElecDensAllAtomCenEnergyCreator.hh |
| Declaration for the class that connects ElecDensAllAtomCenEnergy with the ScoringManager. | |
| file | ElecDensAtomwiseEnergy.cc |
| elec_dens_atomwise scoring method implementation | |
| file | ElecDensAtomwiseEnergy.fwd.hh |
| file | ElecDensAtomwiseEnergy.hh |
| Declaration for elec_dens_atomwise scoring method. | |
| file | ElecDensAtomwiseEnergyCreator.hh |
| file | ElecDensCenEnergy.cc |
| file | ElecDensCenEnergy.hh |
| file | ElecDensCenEnergyCreator.hh |
| Declaration for the class that connects ElecDensCenEnergy with the ScoringManager. | |
| file | ElecDensEnergy.cc |
| file | ElecDensEnergy.hh |
| file | ElecDensEnergyCreator.hh |
| Declaration for the class that connects ElecDensEnergy with the ScoringManager. | |
| file | EnvEnergy.cc |
| file | EnvEnergy.hh |
| EnergyMethod that evaluates env and cbeta energies. | |
| file | EnvEnergyCreator.hh |
| Declaration for the class that connects EnvEnergy with the ScoringManager. | |
| file | EnvSmoothEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | EnvSmoothEnergy.fwd.hh |
| file | EnvSmoothEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | EnvSmoothEnergyCreator.hh |
| Declaration for the class that connects EnvSmoothEnergy with the ScoringManager. | |
| file | ExactOccludedHbondSolEnergy.cc |
| file | ExactOccludedHbondSolEnergy.fwd.hh |
| file | ExactOccludedHbondSolEnergy.hh |
| file | ExactOccludedHbondSolEnergyCreator.hh |
| Declaration for the class that connects ExactOccludedHbondSolEnergy with the ScoringManager. | |
| file | FA_ElecEnergyAroAll.cc |
| Electrostatics energy method for aromatic side chain (stand-in for pi/pi interactions). | |
| file | FA_ElecEnergyAroAll.fwd.hh |
| Electrostatics for RNA class forward declaration. | |
| file | FA_ElecEnergyAroAll.hh |
| file | FA_ElecEnergyAroAllCreator.hh |
| Declaration for the class that connects FA_ElecEnergyAroAll with the ScoringManager. | |
| file | FA_ElecEnergyAroAro.cc |
| Electrostatics energy method for aromatic side chain (stand-in for pi/pi interactions). | |
| file | FA_ElecEnergyAroAro.fwd.hh |
| Electrostatics for RNA class forward declaration. | |
| file | FA_ElecEnergyAroAro.hh |
| file | FA_ElecEnergyAroAroCreator.hh |
| Declaration for the class that connects FA_ElecEnergyAroAro with the ScoringManager. | |
| file | FA_GrpElecEnergy.cc |
| file | FA_GrpElecEnergy.fwd.hh |
| file | FA_GrpElecEnergy.hh |
| file | FA_GrpElecEnergyCreator.hh |
| Declaration for the class that connects FA_ElecEnergy with the ScoringManager. | |
| file | Fa_MbenvEnergy.cc |
| file | Fa_MbenvEnergy.fwd.hh |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method forward declaration. | |
| file | Fa_MbenvEnergy.hh |
| file | Fa_MbenvEnergyCreator.hh |
| Declaration for the class that connects Fa_MbenvEnergy with the ScoringManager. | |
| file | Fa_MbsolvEnergy.cc |
| file | Fa_MbsolvEnergy.fwd.hh |
| file | Fa_MbsolvEnergy.hh |
| LK Solvation using hemisphere culling class forward declaration. | |
| file | Fa_MbsolvEnergyCreator.hh |
| Declaration for the class that connects Fa_MbsolvEnergy with the ScoringManager. | |
| file | FACTSEnergy.cc |
| file | FACTSEnergy.fwd.hh |
| file | FACTSEnergy.hh |
| file | FACTSEnergyCreator.hh |
| file | FaMPAsymEzCBEnergy.cc |
| file | FaMPAsymEzCBEnergy.fwd.hh |
| file | FaMPAsymEzCBEnergy.hh |
| file | FaMPAsymEzCBEnergyCreator.hh |
| file | FaMPAsymEzCGEnergy.cc |
| file | FaMPAsymEzCGEnergy.fwd.hh |
| file | FaMPAsymEzCGEnergy.hh |
| file | FaMPAsymEzCGEnergyCreator.hh |
| file | FaMPEnvEnergy.cc |
| file | FaMPEnvEnergy.fwd.hh |
| file | FaMPEnvEnergy.hh |
| LK-Type Membrane Environment Energy. | |
| file | FaMPEnvEnergyCreator.hh |
| file | FaMPEnvSmoothEnergy.cc |
| file | FaMPEnvSmoothEnergy.fwd.hh |
| file | FaMPEnvSmoothEnergy.hh |
| Fullatom Smoothed Membrane Environment Energy. | |
| file | FaMPEnvSmoothEnergyCreator.hh |
| file | FaMPSolvEnergy.cc |
| LK-Type Membrane Solvation Energy. | |
| file | FaMPSolvEnergy.fwd.hh |
| file | FaMPSolvEnergy.hh |
| LK-Type Membrane Solvation Energy. | |
| file | FaMPSolvEnergyCreator.hh |
| file | FastDensEnergy.cc |
| file | FastDensEnergy.hh |
| file | FastDensEnergyCreator.hh |
| Declaration for the class that connects FastDensEnergy with the ScoringManager. | |
| file | FastSAXSEnergy.cc |
| file | FastSAXSEnergy.hh |
| file | FastSAXSEnergyCreator.hh |
| Energy Creator for FastSAX scoring of Stovgaard et al (BMC Bioinf. 2010) | |
| file | FiberDiffractionEnergy.cc |
| file | FiberDiffractionEnergy.hh |
| file | FiberDiffractionEnergyCreator.hh |
| Energy Creator for FiberDiffractionEnergy. | |
| file | FiberDiffractionEnergyDens.cc |
| file | FiberDiffractionEnergyDens.hh |
| file | FiberDiffractionEnergyDensCreator.hh |
| Energy Creator for FiberDiffractionEnergyDens. | |
| file | FiberDiffractionEnergyGpu.cc |
| file | FiberDiffractionEnergyGpu.hh |
| file | FiberDiffractionEnergyGpuCreator.hh |
| file | FreeDOF_Energy.cc |
| FreeMoiety energy method implementation. | |
| file | FreeDOF_Energy.fwd.hh |
| FreeMoiety energy method forward declaration. | |
| file | FreeDOF_Energy.hh |
| Score function class. | |
| file | FreeDOF_EnergyCreator.hh |
| Declaration for the class that connects FreeDOF_Energy with the ScoringManager. | |
| file | FullatomDisulfideEnergy.cc |
| file | FullatomDisulfideEnergy.fwd.hh |
| file | FullatomDisulfideEnergy.hh |
| file | FullatomDisulfideEnergyCreator.hh |
| Declaration for the class that connects FullatomDisulfideEnergy with the ScoringManager. | |
| file | GaussianOverlapEnergy.cc |
| Gaussian Overlap Energy. | |
| file | GaussianOverlapEnergy.fwd.hh |
| golap | |
| file | GaussianOverlapEnergy.hh |
| energy of overlaps | |
| file | GaussianOverlapEnergyCreator.hh |
| Declaration for the class that connects GaussianOverlapEnergy with the ScoringManager. | |
| file | GenBornEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | GenBornEnergy.fwd.hh |
| file | GenBornEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | GenBornEnergyCreator.hh |
| Declaration for the class that connects GenBornEnergy with the ScoringManager. | |
| file | GenericBondedEnergy.cc |
| A "generic" (atom-type-only-based) torsional potential. | |
| file | GenericBondedEnergy.fwd.hh |
| file | GenericBondedEnergy.hh |
| A "generic" (atom-type-only-based) torsional potential. | |
| file | GenericBondedEnergyCreator.hh |
| Declaration for the class that connects GenericBondedEnergy with the ScoringManager. | |
| file | GoapEnergy.cc |
| C++ implementaion of GOAP(Generalized Orientation-dependent, All-atom statistical Potential) by Zhou H & Skolnick J, Biophys J 2011, 101(8):2043-52. | |
| file | GoapEnergy.fwd.hh |
| file | GoapEnergy.hh |
| file | GoapEnergyCreator.hh |
| file | HackAroEnergy.cc |
| file | HackAroEnergy.fwd.hh |
| file | HackAroEnergy.hh |
| file | HackAroEnergyCreator.hh |
| Declaration for the class that connects HackAroEnergy with the ScoringManager. | |
| file | HolesEnergy.cc |
| file | HolesEnergy.hh |
| file | HolesEnergyCreator.hh |
| Declaration for the class that connects HolesEnergy with the ScoringManager. | |
| file | HRF_MSLabelingEnergy.cc |
| file | HRF_MSLabelingEnergy.fwd.hh |
| file | HRF_MSLabelingEnergy.hh |
| energy term use for scoring predicted HRF_MSLabeling | |
| file | HRF_MSLabelingEnergyCreator.hh |
| Declaration for the class that connects HRF_MSLabelingEnergy with the ScoringManager. | |
| file | HRFDynamicsEnergy.cc |
| Score term calculated based upon neighbor count agreement with hydroxyl radical footprinting labeling data. In a manuscript to be published in 2020. | |
| file | HRFDynamicsEnergy.fwd.hh |
| file | HRFDynamicsEnergy.hh |
| Score term based off mass spectrometry HRF data. | |
| file | HRFDynamicsEnergyCreator.hh |
| Declaration for the class that connects HRFDynamicsEnergy with the ScoringManager. | |
| file | HybridVDW_Energy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | HybridVDW_Energy.fwd.hh |
| file | HybridVDW_Energy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | HybridVDW_EnergyCreator.hh |
| Declaration for the class that connects HybridVDW_Energy with the ScoringManager. | |
| file | HydroxylTorsionEnergy.cc |
| file | HydroxylTorsionEnergy.fwd.hh |
| file | HydroxylTorsionEnergy.hh |
| Term for proton_chi on Ser/Thr/Tyr residues. | |
| file | HydroxylTorsionEnergyCreator.hh |
| file | ImplicitMembraneCoulomb.cc |
| Minimal class for computing the depth- and membrane-dependent electrostatics energy. | |
| file | ImplicitMembraneCoulomb.fwd.hh |
| Minimal class for computing the depth- and membrane-dependent electrostatics energy. | |
| file | ImplicitMembraneCoulomb.hh |
| Minimal class for computing the depth- and membrane-dependent electrostatics energy. | |
| file | ImplicitMembraneElecEnergy.cc |
| Energy method for computing the depth- and membrane-dependent electrostatics energy. | |
| file | ImplicitMembraneElecEnergy.fwd.hh |
| file | ImplicitMembraneElecEnergy.hh |
| Energy method for computing the depth- and membrane-dependent electrostatics energy. | |
| file | ImplicitMembraneElecEnergyCreator.hh |
| file | IntermolEnergy.cc |
| Cost of bringing two chains together. | |
| file | IntermolEnergy.hh |
| Radius of gyration score for METHODS, to match Rosetta++. | |
| file | IntermolEnergyCreator.hh |
| Declaration for the class that connects IntermolEnergy with the ScoringManager. | |
| file | LinearBranchEnergy.cc |
| Atom pair energy functions. | |
| file | LinearBranchEnergy.fwd.hh |
| file | LinearBranchEnergy.hh |
| file | LinearBranchEnergyCreator.cc |
| LinearBranchEnergyCreator implementation. | |
| file | LinearBranchEnergyCreator.hh |
| Declaration for the class that connects LinearBranchEnergy with the ScoringManager. | |
| file | LinearChainbreakEnergy.cc |
| Atom pair energy functions. | |
| file | LinearChainbreakEnergy.fwd.hh |
| file | LinearChainbreakEnergy.hh |
| file | LinearChainbreakEnergyCreator.cc |
| LinearChainbreakEnergyCreator implementation. | |
| file | LinearChainbreakEnergyCreator.hh |
| Declaration for the class that connects LinearChainbreakEnergy with the ScoringManager. | |
| file | LK_hack.cc |
| file | LK_hack.fwd.hh |
| file | LK_hack.hh |
| LK Solvation using hemisphere culling class declaration. | |
| file | LK_hackCreator.hh |
| Declaration for the class that connects LK_hack with the ScoringManager. | |
| file | LK_PolarNonPolarEnergy.cc |
| file | LK_PolarNonPolarEnergy.fwd.hh |
| file | LK_PolarNonPolarEnergy.hh |
| LK Solvation using hemisphere culling class forward declaration. | |
| file | LK_PolarNonPolarEnergyCreator.hh |
| Declaration for the class that connects LK_PolarNonPolarEnergy with the ScoringManager. | |
| file | LoopCloseEnergy.cc |
| Cost of bringing two chains together. | |
| file | LoopCloseEnergy.hh |
| file | LoopCloseEnergyCreator.hh |
| Declaration for the class that connects LoopCloseEnergy with the ScoringManager. | |
| file | MembraneCbetaEnergy.cc |
| file | MembraneCbetaEnergy.fwd.hh |
| file | MembraneCbetaEnergy.hh |
| Membrane Environment Cbeta Energy. | |
| file | MembraneCbetaEnergyCreator.hh |
| Declaration for the class that connects MembraneCbetaEnergy with the ScoringManager. | |
| file | MembraneCenPairEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | MembraneCenPairEnergy.fwd.hh |
| file | MembraneCenPairEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | MembraneCenPairEnergyCreator.hh |
| Declaration for the class that connects MembraneCenPairEnergy with the ScoringManager. | |
| file | MembraneEnvEnergy.cc |
| file | MembraneEnvEnergy.fwd.hh |
| file | MembraneEnvEnergy.hh |
| Membrane Environment Cbeta Energy. | |
| file | MembraneEnvEnergyCreator.hh |
| Declaration for the class that connects MembraneEnvEnergy with the ScoringManager. | |
| file | MembraneEnvPenalties.cc |
| file | MembraneEnvPenalties.fwd.hh |
| file | MembraneEnvPenalties.hh |
| RMS Energy function. Used to optimize the RMSD between two structures. | |
| file | MembraneEnvPenaltiesCreator.hh |
| Declaration for the class that connects MembraneEnvPenalties with the ScoringManager. | |
| file | MembraneEnvSmoothEnergy.cc |
| Statistically derived smooth membrane environment potential class implementation. | |
| file | MembraneEnvSmoothEnergy.fwd.hh |
| file | MembraneEnvSmoothEnergy.hh |
| Statistically derived smooth membrane environment potential class declaration. | |
| file | MembraneEnvSmoothEnergyCreator.hh |
| Declaration for the class that connects MembraneEnvSmoothEnergy with the ScoringManager. | |
| file | MembraneLipo.cc |
| file | MembraneLipo.fwd.hh |
| file | MembraneLipo.hh |
| RMS Energy function. Used to optimize the RMSD between two structures. | |
| file | MembraneLipoCreator.hh |
| Declaration for the class that connects MembraneLipo with the ScoringManager. | |
| file | MgEnergy.cc |
| file | MgEnergy.fwd.hh |
| file | MgEnergy.hh |
| file | MgEnergyCreator.hh |
| Declaration for the class that connects MgEnergy with the ScoringManager. | |
| file | MHCEpitopeEnergy.cc |
| file | MHCEpitopeEnergy.fwd.hh |
| file | MHCEpitopeEnergy.hh |
| file | MHCEpitopeEnergyCreator.hh |
| file | MissingEnergy.cc |
| Cost of failing to close loops. | |
| file | MissingEnergy.hh |
| Cost of failing to close loops. | |
| file | MissingEnergyCreator.hh |
| Declaration for the class that connects MissingEnergy with the ScoringManager. | |
| file | MotifDockEnergy.cc |
| Adaptation of Motif score for Docking. | |
| file | MotifDockEnergy.fwd.hh |
| file | MotifDockEnergy.hh |
| Adaptation of Motif score for Docking. | |
| file | MotifDockEnergyCreator.hh |
| Adaptation of Motif score for Docking. | |
| file | MPCBetaEnergy.cc |
| Membrane Environemnt CBeta Energy. | |
| file | MPCBetaEnergy.fwd.hh |
| file | MPCBetaEnergy.hh |
| file | MPCBetaEnergyCreator.hh |
| file | MPEnvEnergy.cc |
| Membrane Environemnt Energy. | |
| file | MPEnvEnergy.fwd.hh |
| Membrane Environemnt Energy. | |
| file | MPEnvEnergy.hh |
| Membrane Environemnt Energy. | |
| file | MPEnvEnergyCreator.hh |
| file | MPHelicalityEnergy.cc |
| Fullatom and centroid level smooth membrane non-helicality penalty. | |
| file | MPHelicalityEnergy.fwd.hh |
| file | MPHelicalityEnergy.hh |
| Fullatom and centroid level smooth membrane non-helicality penalty. | |
| file | MPHelicalityEnergyCreator.hh |
| file | MPPairEnergy.cc |
| Membrane Residue Pair Energy Term. | |
| file | MPPairEnergy.fwd.hh |
| file | MPPairEnergy.hh |
| Membrane Residue Pair Energy Term. | |
| file | MPPairEnergyCreator.hh |
| file | MPResidueLipophilicityEnergy.cc |
| Fullatom Smoothed Membrane Environment Energy. | |
| file | MPResidueLipophilicityEnergy.fwd.hh |
| file | MPResidueLipophilicityEnergy.hh |
| Fullatom Smoothed Membrane Environment Energy. | |
| file | MPResidueLipophilicityEnergyCreator.hh |
| file | MPSpanAngleEnergy.cc |
| penalize spans with unnatural dG of insertion | |
| file | MPSpanAngleEnergy.hh |
| file | MPSpanAngleEnergyCreator.hh |
| file | MPSpanInsertionEnergy.cc |
| penalize spans with unnatural dG of insertion | |
| file | MPSpanInsertionEnergy.hh |
| file | MPSpanInsertionEnergyCreator.hh |
| file | MultipoleElecEnergy.cc |
| Fixed multipole electrostatics using Jay Ponder's approach in Tinker/Amoeba. | |
| file | MultipoleElecEnergy.fwd.hh |
| file | MultipoleElecEnergy.hh |
| Fixed multipole electrostatics using Jay Ponder's approach in Tinker/Amoeba. | |
| file | MultipoleElecEnergyCreator.hh |
| Declaration for the class that connects MultipoleElecEnergy with the ScoringManager. | |
| file | NetChargeEnergy.cc |
| file | NetChargeEnergy.fwd.hh |
| file | NetChargeEnergy.hh |
| file | NetChargeEnergyCreator.hh |
| file | non_scorefxn_exact_model.cc |
| file | non_scorefxn_exact_model.hh |
| file | NPDHBondEnergy.cc |
| file | NPDHBondEnergy.fwd.hh |
| file | NPDHBondEnergy.hh |
| file | NPDHBondEnergyCreator.hh |
| Declaration for the class that connects NPDHBondEnergy with the ScoringManager. | |
| file | OccludedHbondSolEnergy.cc |
| file | OccludedHbondSolEnergy.fwd.hh |
| file | OccludedHbondSolEnergy.hh |
| file | OccludedHbondSolEnergy_onebody.cc |
| file | OccludedHbondSolEnergy_onebody.fwd.hh |
| file | OccludedHbondSolEnergy_onebody.hh |
| file | OccludedHbondSolEnergy_onebodyCreator.hh |
| Declaration for the class that connects OccludedHbondSolEnergy_onebody with the ScoringManager. | |
| file | OccludedHbondSolEnergyCreator.hh |
| Declaration for the class that connects OccludedHbondSolEnergy with the ScoringManager. | |
| file | OmegaTetherEnergy.cc |
| OmegaTether energy method class implementation. | |
| file | OmegaTetherEnergy.fwd.hh |
| OmegaTether energy method class forward declaration. | |
| file | OmegaTetherEnergy.hh |
| OmegaTether energy method class declaration. | |
| file | OmegaTetherEnergyCreator.hh |
| Declaration for the class that connects OmegaTetherEnergy with the ScoringManager. | |
| file | OtherPoseEnergy.cc |
| Way to handle scoring of a collection of poses, allowing shift of focus between poses. | |
| file | OtherPoseEnergy.fwd.hh |
| file | OtherPoseEnergy.hh |
| file | OtherPoseEnergyCreator.hh |
| Declaration for the class that connects OtherPoseEnergy with the ScoringManager. | |
| file | P_AA_Energy.cc |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method implementation. | |
| file | P_AA_Energy.fwd.hh |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method forward declaration. | |
| file | P_AA_Energy.hh |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method declaration. | |
| file | P_AA_EnergyCreator.hh |
| Declaration for the class that connects P_AA_Energy with the ScoringManager. | |
| file | P_AA_pp_Energy.cc |
| Probability of observing an amino acid, given its phi/psi energy method declaration. | |
| file | P_AA_pp_Energy.fwd.hh |
| file | P_AA_pp_Energy.hh |
| Probability of observing an amino acid, given its phi/psi energy method forward declaration. | |
| file | P_AA_pp_EnergyCreator.hh |
| Declaration for the class that connects P_AA_pp_Energy with the ScoringManager. | |
| file | P_AA_ss_Energy.cc |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method implementation. | |
| file | P_AA_ss_Energy.fwd.hh |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method forward declaration. | |
| file | P_AA_ss_Energy.hh |
| Probability of observing an amino acid (NOT conditional on phi/psi), energy method declaration. | |
| file | P_AA_ss_EnergyCreator.hh |
| Declaration for the class that connects P_AA_ss_Energy with the ScoringManager. | |
| file | PackStatEnergy.cc |
| Radius of gyration energy function definition. Returns -1 * RG for a given Pose. | |
| file | PackStatEnergy.hh |
| file | PackStatEnergyCreator.hh |
| Declaration for the class that connects PackStatEnergy with the ScoringManager. | |
| file | PairEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | PairEnergy.fwd.hh |
| Statistically derived rotamer pair potential class forward declaration. | |
| file | PairEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | PairEnergyCreator.hh |
| Declaration for the class that connects PairEnergy with the ScoringManager. | |
| file | PeptideBondEnergy.cc |
| file | PeptideBondEnergy.fwd.hh |
| file | PeptideBondEnergy.hh |
| scoring method that defines ideal bond lengths between carbonyl carbon of residue N and nitrogen of residue N+1. | |
| file | PeptideBondEnergyCreator.hh |
| Declaration for the class that connects PeptideBondEnergy with the ScoringManager. | |
| file | pHEnergy.cc |
| Energy due to ionization state of the residue at a particular pH (see pHEnergy.hh for a detailed description) | |
| file | pHEnergy.fwd.hh |
| pH energy forward declaration | |
| file | pHEnergy.hh |
| file | pHEnergyCreator.hh |
| Declaration for the class that connects pHEnergy with the ScoringManager. | |
| file | PointWaterEnergy.cc |
| file | PointWaterEnergy.fwd.hh |
| file | PointWaterEnergy.hh |
| Statistical point water energy function. | |
| file | PointWaterEnergyCreator.hh |
| file | PoissonBoltzmannEnergy.cc |
| Ramachandran energy method class implementation. | |
| file | PoissonBoltzmannEnergy.fwd.hh |
| Ramachandran energy method class forward declaration. | |
| file | PoissonBoltzmannEnergy.hh |
| Membrane Environment Cbeta Energy. | |
| file | PoissonBoltzmannEnergyCreator.hh |
| file | ProClosureEnergy.cc |
| Proline ring closure energy method class implementation. | |
| file | ProClosureEnergy.fwd.hh |
| Proline ring closure energy method class forward declaration. | |
| file | ProClosureEnergy.hh |
| Proline ring closure energy method class declaration. | |
| file | ProClosureEnergyCreator.hh |
| Declaration for the class that connects ProClosureEnergy with the ScoringManager. | |
| file | ProQ_Energy.cc |
| ProQ energy function definition. | |
| file | ProQ_Energy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | ProQ_EnergyCreator.hh |
| Declaration for the class that connects ProQ_Energy with the ScoringManager. | |
| file | PyEnergy.hh |
| Various Energy classes for subclassing in PyRosetta. | |
| file | RamachandranEnergy.cc |
| Ramachandran energy method class implementation. | |
| file | RamachandranEnergy.fwd.hh |
| Ramachandran energy method class forward declaration. | |
| file | RamachandranEnergy.hh |
| Ramachandran energy method class declaration. | |
| file | RamachandranEnergy2B.cc |
| file | RamachandranEnergy2B.fwd.hh |
| file | RamachandranEnergy2B.hh |
| Ramachandran energy method class declaration. | |
| file | RamachandranEnergy2BCreator.hh |
| Declaration for the class that connects RamachandranEnergy2B with the ScoringManager. | |
| file | RamachandranEnergyCreator.hh |
| Declaration for the class that connects RamachandranEnergy with the ScoringManager. | |
| file | RamaPreProEnergy.cc |
| A variation on the Ramachandran scorefunction that has separate probability tables for residues that precede prolines. | |
| file | RamaPreProEnergy.fwd.hh |
| file | RamaPreProEnergy.hh |
| A variation on the Ramachandran scorefunction that has separate probability tables for residues that precede prolines. | |
| file | RamaPreProEnergyCreator.hh |
| Declaration for the class that connects RamaPreProEnergy with the ScoringManager. | |
| file | ReferenceEnergy.cc |
| file | ReferenceEnergy.fwd.hh |
| Reference energy method forward declaration. | |
| file | ReferenceEnergy.hh |
| Reference energy method implementation. | |
| file | ReferenceEnergyCreator.hh |
| Declaration for the class that connects ReferenceEnergy with the ScoringManager. | |
| file | ReferenceEnergyNoncanonical.cc |
| Reference energy method implementation. | |
| file | ReferenceEnergyNoncanonical.fwd.hh |
| Reference energy method forward declaration. | |
| file | ReferenceEnergyNoncanonical.hh |
| Reference energy method declaration. | |
| file | ReferenceEnergyNoncanonicalCreator.hh |
| Declaration for the class that connects ReferenceEnergyNoncanonical with the ScoringManager. | |
| file | ResidualDipolarCouplingEnergy.cc |
| RDC energy - comparing experimental RDC values to calculated values. | |
| file | ResidualDipolarCouplingEnergy.fwd.hh |
| file | ResidualDipolarCouplingEnergy.hh |
| RDC energy - comparing experimental RDC values to calculated values. | |
| file | ResidualDipolarCouplingEnergy_Rohl.cc |
| RDC energy - comparing experimental RDC values to calculated values. | |
| file | ResidualDipolarCouplingEnergy_Rohl.hh |
| file | ResidualDipolarCouplingEnergy_RohlCreator.hh |
| Declaration for the class that connects ResidualDipolarCouplingEnergy_Rohl with the ScoringManager. | |
| file | ResidualDipolarCouplingEnergyCreator.hh |
| Declaration for the class that connects ResidualDipolarCouplingEnergy with the ScoringManager. | |
| file | RG_Energy_Fast.cc |
| Radius of gyration energy function definition. | |
| file | RG_Energy_Fast.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | RG_Energy_FastCreator.hh |
| Declaration for the class that connects RG_Energy_Fast with the ScoringManager. | |
| file | RG_Energy_RNA.cc |
| file | RG_Energy_RNA.hh |
| file | RG_Energy_RNACreator.hh |
| Declaration for the class that connects RG_Energy_RNA with the ScoringManager. | |
| file | RG_LocalEnergy.cc |
| Radius of gyration for local region. | |
| file | RG_LocalEnergy.hh |
| only calculates RG the length of the repeat. Uses code from RG_Energy_fast | |
| file | RG_LocalEnergyCreator.hh |
| file | RingClosureEnergy.cc |
| Noncanonical ring closure energy method class implementation. | |
| file | RingClosureEnergy.fwd.hh |
| Noncanonical ring closure energy method class forward declaration. | |
| file | RingClosureEnergy.hh |
| Noncanonical ring closure energy method class headers. | |
| file | RingClosureEnergyCreator.hh |
| Declaration for the class that connects RingClosureEnergy with the ScoringManager. | |
| file | RMS_Energy.cc |
| RMS Energy function. Used to optimize the RMSD between two structures. | |
| file | RMS_Energy.hh |
| RMS Energy function. Used to optimize the RMSD between two structures. | |
| file | RMS_EnergyCreator.hh |
| Declaration for the class that connects RMS_Energy with the ScoringManager. | |
| file | RNA_BulgeEnergy.cc |
| file | RNA_BulgeEnergy.fwd.hh |
| file | RNA_BulgeEnergy.hh |
| file | RNA_BulgeEnergyCreator.hh |
| Declaration for the class that connects RNA_BulgeEnergy with the ScoringManager. | |
| file | RNA_ChemicalMappingEnergy.cc |
| file | RNA_ChemicalMappingEnergy.fwd.hh |
| file | RNA_ChemicalMappingEnergy.hh |
| file | RNA_ChemicalMappingEnergyCreator.hh |
| file | RNA_ChemicalShiftEnergy.cc |
| Energy score based on the agreement between experimentally determined and theoretically calculated NMR chemical_shift. | |
| file | RNA_ChemicalShiftEnergy.hh |
| Energy score based on the agreement between experimentally determined and theoretically calculated NMR chemical_shift. | |
| file | RNA_ChemicalShiftEnergyCreator.hh |
| file | RNA_CoarseDistEnergy.cc |
| Score two-body energies in coarse RNA poses between P, S, and CEN using a statistical potential. | |
| file | RNA_CoarseDistEnergy.fwd.hh |
| Score two-body energies in coarse RNA poses between P, S, and CEN using a statistical potential. | |
| file | RNA_CoarseDistEnergy.hh |
| Score two-body energies in coarse RNA poses between P, S, and CEN using a statistical potential. | |
| file | RNA_CoarseDistEnergyCreator.hh |
| Score two-body energies in coarse RNA poses between P, S, and CEN using a statistical potential. | |
| file | RNA_DataBackboneEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | RNA_DataBackboneEnergy.fwd.hh |
| file | RNA_DataBackboneEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | RNA_DataBackboneEnergyCreator.hh |
| file | RNA_FA_ElecEnergy.cc |
| Electrostatics energy method for RNA class implementation. | |
| file | RNA_FA_ElecEnergy.fwd.hh |
| Electrostatics for RNA class forward declaration. | |
| file | RNA_FA_ElecEnergy.hh |
| file | RNA_FA_ElecEnergyCreator.hh |
| Declaration for the class that connects RNA_FA_ElecEnergy with the ScoringManager. | |
| file | RNA_FullAtomStackingEnergy.cc |
| file | RNA_FullAtomStackingEnergy.fwd.hh |
| file | RNA_FullAtomStackingEnergy.hh |
| file | RNA_FullAtomStackingEnergyCreator.hh |
| Declaration for the class that connects RNA_FullAtomStackingEnergy with the ScoringManager. | |
| file | RNA_FullAtomVDW_BasePhosphate.cc |
| file | RNA_FullAtomVDW_BasePhosphate.fwd.hh |
| file | RNA_FullAtomVDW_BasePhosphate.hh |
| FullAtom VDW energy between the base and phosphate group in the same (intra) nucleotide. | |
| file | RNA_FullAtomVDW_BasePhosphateCreator.fwd.hh |
| file | RNA_FullAtomVDW_BasePhosphateCreator.hh |
| file | RNA_JR_SuiteEnergy.cc |
| file | RNA_JR_SuiteEnergy.fwd.hh |
| file | RNA_JR_SuiteEnergy.hh |
| file | RNA_JR_SuiteEnergyCreator.hh |
| file | RNA_LJ_BaseEnergy.cc |
| file | RNA_LJ_BaseEnergy.fwd.hh |
| file | RNA_LJ_BaseEnergy.hh |
| file | RNA_LJ_BaseEnergyCreator.hh |
| Declaration for the class that connects RNA_LJ_BaseEnergy with the ScoringManager. | |
| file | RNA_MgPointEnergy.cc |
| Statistically derived Mg(2+) binding potential for RNA. | |
| file | RNA_MgPointEnergy.hh |
| Statistically derived Mg(2+) binding potential for RNA. | |
| file | RNA_MgPointEnergyCreator.hh |
| Declaration for the class that connects RNA_MgPointEnergy with the ScoringManager. | |
| file | RNA_PairwiseLowResolutionEnergy.cc |
| file | RNA_PairwiseLowResolutionEnergy.fwd.hh |
| file | RNA_PairwiseLowResolutionEnergy.hh |
| file | RNA_PairwiseLowResolutionEnergyCreator.hh |
| Declaration for the class that connects RNA_PairwiseLowResolutionEnergy with the ScoringManager. | |
| file | RNA_PartitionEnergy.cc |
| file | RNA_PartitionEnergy.hh |
| file | RNA_PartitionEnergyCreator.hh |
| Declaration for the class that connects RNA_PartitionEnergy with the ScoringManager. | |
| file | RNA_StubCoordinateEnergy.cc |
| file | RNA_StubCoordinateEnergy.fwd.hh |
| file | RNA_StubCoordinateEnergy.hh |
| file | RNA_StubCoordinateEnergyCreator.hh |
| Declaration for the class that connects RNA_StubCoordinateEnergy with the ScoringManager. | |
| file | RNA_SugarCloseEnergy.cc |
| file | RNA_SugarCloseEnergy.fwd.hh |
| file | RNA_SugarCloseEnergy.hh |
| file | RNA_SugarCloseEnergyCreator.hh |
| Declaration for the class that connects RNA_SugarCloseEnergy with the ScoringManager. | |
| file | RNA_SuiteEnergy.cc |
| file | RNA_SuiteEnergy.fwd.hh |
| file | RNA_SuiteEnergy.hh |
| file | RNA_SuiteEnergyCreator.hh |
| Declaration for the class that connects RNA_ChemicalMappingEnergy with the ScoringManager. | |
| file | RNA_TorsionEnergy.cc |
| file | RNA_TorsionEnergy.fwd.hh |
| file | RNA_TorsionEnergy.hh |
| file | RNA_TorsionEnergyCreator.hh |
| Declaration for the class that connects RNA_TorsionEnergy with the ScoringManager. | |
| file | RNA_VDW_Energy.cc |
| file | RNA_VDW_Energy.fwd.hh |
| file | RNA_VDW_Energy.hh |
| file | RNA_VDW_EnergyCreator.hh |
| Declaration for the class that connects RNA_VDW_Energy with the ScoringManager. | |
| file | RNP_LowResEnergy.cc |
| file | RNP_LowResEnergy.fwd.hh |
| file | RNP_LowResEnergy.hh |
| file | RNP_LowResEnergyCreator.hh |
| Energy method creator for the low resolution RNA/protein potential. | |
| file | RNP_LowResPairDistEnergy.cc |
| file | RNP_LowResPairDistEnergy.fwd.hh |
| file | RNP_LowResPairDistEnergy.hh |
| file | RNP_LowResPairDistEnergyCreator.hh |
| Energy method creator for the low resolution RNA/protein potential. | |
| file | RNP_LowResStackEnergy.cc |
| file | RNP_LowResStackEnergy.fwd.hh |
| file | RNP_LowResStackEnergy.hh |
| file | RNP_LowResStackEnergyCreator.hh |
| Energy method creator for the low resolution RNA/protein potential. | |
| file | SA_Energy.cc |
| sa energy function definition. | |
| file | SA_Energy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | SA_EnergyCreator.hh |
| file | SASAEnergy.cc |
| Power Diagram-derived solvent-accessible surface area energy. | |
| file | SASAEnergy.fwd.hh |
| Power Diagram-derived solvent-accessible surface area energy. | |
| file | SASAEnergy.hh |
| Power Diagram-derived solvent-accessible surface area energy. | |
| file | SASAEnergyCreator.hh |
| Declaration for the class that connects SASAEnergy with the ScoringManager. | |
| file | SAXSEnergy.cc |
| FastSAX scoring of Stovgaard et al (BMC Bioinf. 2010) | |
| file | SAXSEnergy.hh |
| file | SAXSEnergyCEN.hh |
| file | SAXSEnergyCreator.hh |
| Declaration for the class that connects SAXSEnergyCreator with the ScoringManager. | |
| file | SAXSEnergyCreatorCEN.cc |
| file | SAXSEnergyCreatorCEN.hh |
| Declaration for the class that connects SAXSEnergyCreatorCEN with the ScoringManager. | |
| file | SAXSEnergyCreatorFA.cc |
| file | SAXSEnergyCreatorFA.hh |
| Declaration for the class that connects SAXSEnergyCreatorFA with the ScoringManager. | |
| file | SAXSEnergyFA.hh |
| file | SecondaryStructureEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | SecondaryStructureEnergy.fwd.hh |
| file | SecondaryStructureEnergy.hh |
| Statistically derived rotamer pair potential class declaration. | |
| file | SecondaryStructureEnergyCreator.hh |
| Declaration for the class that connects SecondaryStructureEnergy with the ScoringManager. | |
| file | SequenceDependentRefEnergy.cc |
| file | SequenceDependentRefEnergy.fwd.hh |
| Reference energy method forward declaration. | |
| file | SequenceDependentRefEnergy.hh |
| Reference energy method implementation. | |
| file | SequenceDependentRefEnergyCreator.hh |
| Declaration for the class that connects SequenceDependentRefEnergy with the ScoringManager. | |
| file | SmoothCenPairEnergy.cc |
| Smooth, differentiable, version of cenpair. | |
| file | SmoothCenPairEnergy.hh |
| file | SmoothCenPairEnergyCreator.hh |
| file | SmoothEnvEnergy.cc |
| Smooth, differentiable, version of centroid env. | |
| file | SmoothEnvEnergy.hh |
| file | SmoothEnvEnergyCreator.hh |
| file | SplitUnfoldedTwoBodyEnergy.cc |
| Split unfolded energy two body energy method. | |
| file | SplitUnfoldedTwoBodyEnergy.fwd.hh |
| Forward header for energy method for the two body portion of the split unfolded energy. | |
| file | SplitUnfoldedTwoBodyEnergy.hh |
| Header for the split unfolded energy two body component energy method. | |
| file | SplitUnfoldedTwoBodyEnergyCreator.cc |
| file | SplitUnfoldedTwoBodyEnergyCreator.hh |
| Energy creator for the split unfolded two body energy method. | |
| file | SSElementMotifContactEnergy.cc |
| file | SSElementMotifContactEnergy.h |
| file | SSElementMotifContactEnergy.hh |
| Will's motif score to determine how well packed the protein core is. | |
| file | SSElementMotifContactEnergyCreator.hh |
| file | StackElecEnergy.cc |
| file | StackElecEnergy.fwd.hh |
| file | StackElecEnergy.hh |
| file | StackElecEnergyCreator.hh |
| Declaration for the class that connects StackElecEnergy with the ScoringManager. | |
| file | SuckerEnergy.cc |
| Statistically derived rotamer pair potential class implementation. | |
| file | SuckerEnergy.fwd.hh |
| Statistically derived rotamer pair potential class forward declaration. | |
| file | SuckerEnergy.hh |
| energy attracting atoms to sucker atoms | |
| file | SuckerEnergyCreator.hh |
| Declaration for the class that connects SuckerEnergy with the ScoringManager. | |
| file | SugarBackboneEnergy.cc |
| file | SugarBackboneEnergy.fwd.hh |
| Forward declarations for SugarBackboneEnergy. | |
| file | SugarBackboneEnergy.hh |
| Method definitions for SugarBackboneEnergy and SugarBackboneEnergyCreator. | |
| file | SugarBackboneEnergyCreator.hh |
| file | SurfEnergy.cc |
| Packing Score. | |
| file | SurfEnergy.hh |
| Packing Score. | |
| file | SurfEnergyCreator.hh |
| Declaration for the class that connects SurfEnergy with the ScoringManager. | |
| file | SurfVolEnergy.cc |
| Packing Score. | |
| file | SurfVolEnergy.hh |
| file | SurfVolEnergyCreator.hh |
| Declaration for the class that connects SurfVolEnergy with the ScoringManager. | |
| file | SymmetricLigandEnergy.cc |
| Dunbrack energy method implementation. | |
| file | SymmetricLigandEnergy.fwd.hh |
| Dunbrack energy method forward declaration. | |
| file | SymmetricLigandEnergy.hh |
| score for implicit ligand interactions from symmetric geometry | |
| file | SymmetricLigandEnergyCreator.hh |
| file | TNA_SuiteEnergy.cc |
| file | TNA_SuiteEnergy.fwd.hh |
| file | TNA_SuiteEnergy.hh |
| file | TNA_SuiteEnergyCreator.hh |
| Declaration for the class that connects TNA_SuiteEnergy with the ScoringManager. | |
| file | UnfoldedStateEnergy.cc |
| Unfolded state energy method implementation; energies based on eneriges of residues in fragments. | |
| file | UnfoldedStateEnergy.fwd.hh |
| Unfolded state energy method, forward declaration. | |
| file | UnfoldedStateEnergy.hh |
| Unfolded state energy declaration header file. | |
| file | UnfoldedStateEnergyCreator.hh |
| Declaration for the class that connects UnfoldedStateEnergy with the ScoringManager. | |
| file | VdWTinkerEnergy.cc |
| VdW treatment using buffered 14-7 approach in Tinker/Amoeba. | |
| file | VdWTinkerEnergy.fwd.hh |
| file | VdWTinkerEnergy.hh |
| file | VdWTinkerEnergyCreator.hh |
| Declaration for the class that connects VdWTinkerEnergy with the ScoringManager. | |
| file | WaterAdductHBondEnergy.cc |
| file | WaterAdductHBondEnergy.fwd.hh |
| file | WaterAdductHBondEnergy.hh |
| file | WaterAdductHBondEnergyCreator.hh |
| Declaration for the class that connects WaterAdductHBondEnergy with the ScoringManager. | |
| file | WaterAdductIntraEnergy.cc |
| file | WaterAdductIntraEnergy.fwd.hh |
| file | WaterAdductIntraEnergy.hh |
| Energetic offset/cost for placing a water adduct on an amino or nucleic acid. | |
| file | WaterAdductIntraEnergyCreator.hh |
| Declaration for the class that connects WaterAdductIntraEnergy with the ScoringManager. | |
| file | WaterSpecificEnergy.cc |
| file | WaterSpecificEnergy.fwd.hh |
| file | WaterSpecificEnergy.hh |
| Energetic considerations for explicit water molecules,. | |
| file | WaterSpecificEnergyCreator.hh |
| Declaration for the class that connects WaterSpecificEnergy with the ScoringManager. | |
| file | XtalMLEnergy.cc |
| file | XtalMLEnergy.hh |
| file | XtalMLEnergyCreator.hh |
| ML target. | |
| file | YHHPlanarityEnergy.cc |
| file | YHHPlanarityEnergy.fwd.hh |
| file | YHHPlanarityEnergy.hh |
| Term for chi3 on tyrosine residues to prefer the hydrogen lie in the plane of the ring. | |
| file | YHHPlanarityEnergyCreator.hh |