File Format
Overview
Prior knowledge about a protein's structure is conveyed to the NonlocalAbinitio protocol by representing that information hierarchically in a structured text file (often referred to as a pairings file). Conceptually, a pairings file consists of 3 types of entities-- NLGrouping's, NLFragmentGroup's, and NLFragment's. We'll discuss these in reverse order.
NLFragment's contain structural information about a single residue in a protein. This information includes residue number, backbone torsion angles, and the Cartesian coordinates of the residue's Cα atom. Entries are comma-separated and may contain leading and trailing whitespace. Represented in Backus-Naur Form:
<NLFragment> ::= <residue> "," <phi> "," <psi> "," <omega> "," <CA x> "," <CA y> "," <CA z> <EOL>
NLFragment's belonging to the same region (e.g. secondary structure element) are grouped together by listing them consecutively, one per line. When a newline is encountered, a new NLFragmentGroup is started. Conceptually, NLFragmentGroup's play an analogous role to Rosetta's core.fragment.FragData class. The number of NLFragment's in each NLFragmentGroup need not be the same.
In the same manner that NLFragmentGroup's contained multiple NLFragment's, NLGrouping's contain multiple NLFragmentGroup's. Syntactically, NLGrouping's are enclosed within curly braces {}. To motivate their use, consider the following situation. Assume we have a predictive method for identifying contacting regions of a protein.
Example
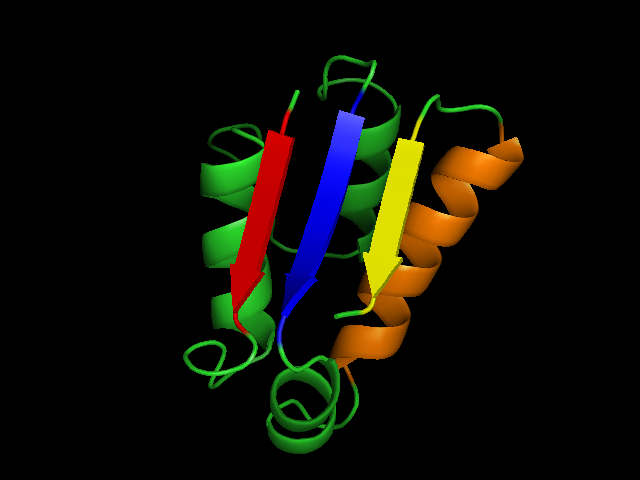
To take a concrete example, consider the file shown below, which defines a single NLGrouping that contains 4 NLFragmentGroup's. The correspondences between NLFragmentGroup's and protein regions are shown below.
- NLFragmentGroup~1~ (residues 2-6) => red
- NLFragmentGroup~2~ (residues 49-54) => blue
- NLFragmentGroup~3~ (residues 66-80) => orange
- NLFragmentGroup~4~ (residues 84-88) => yellow
An example used to illustrate the mapping between segments in the non-local abinitio file format and sections of protein 1a19a.
{
2,-110.466,126.421,174.600,97.539,49.731,-2.920
3,-119.756,145.382,-177.458,96.634,51.368,0.388
4,-132.235,119.796,176.520,95.461,49.639,3.564
5,-117.492,124.130,177.571,93.691,51.271,6.506
6,-107.749,99.134,179.550,92.793,49.314,9.639
49,-143.132,128.827,176.942,94.852,53.324,-6.631
50,-118.006,112.700,-171.815,93.025,50.630,-4.667
51,-105.538,117.641,178.295,92.132,51.602,-1.106
52,-102.343,117.860,179.175,91.170,48.933,1.425
53,-107.322,91.600,179.354,89.445,50.179,4.575
54,-71.162,135.072,176.466,89.341,47.329,7.090
66,-81.450,0.155,-178.645,85.342,62.214,11.149
67,-84.864,-23.372,-174.554,86.194,59.236,8.944
68,-73.413,-31.321,178.657,83.172,59.675,6.682
69,-70.632,-42.954,172.619,84.356,63.044,5.377
70,-54.791,-54.004,-178.833,87.738,61.605,4.420
71,-57.600,-35.842,-179.338,86.085,58.712,2.589
72,-69.881,-27.140,172.866,83.805,61.185,0.815
73,-67.993,-40.198,171.931,86.903,62.991,-0.438
74,-67.543,-40.261,175.771,88.036,59.706,-1.968
75,-63.606,-23.738,-176.963,84.483,59.165,-3.205
76,-85.821,-44.670,174.120,84.733,62.625,-4.760
77,-55.205,-47.212,172.312,88.076,62.176,-6.508
78,-51.331,-48.179,179.400,86.652,59.002,-8.032
79,-56.568,-59.022,-177.892,83.667,61.050,-9.195
80,-53.290,-31.993,168.475,85.700,63.839,-10.789
84,-118.247,139.397,171.264,90.253,54.853,-7.446
85,-118.683,127.693,-173.555,88.952,51.548,-6.102
86,-116.318,111.870,175.979,87.492,51.458,-2.596
87,-100.612,112.101,-179.332,87.116,48.184,-0.701
88,-93.093,77.996,-179.257,85.043,48.589,2.460
}
Code
The data types defined above are located in NLFragment.hh, NLFragmentGroup.hh, and NLGrouping.hh respectively. NLGrouping's can be constructed in one of two ways. They can be read explicitly from file using NonlocalAbinitioReader's static read() method.
#include <protocols/abinitio/NonlocalAbinitioReader.hh>
...
vector1<NLGrouping> groupings;
NonlocalAbinitioReader::read("test.pairings", &groupings);
Alternately, NLGrouping's can be constructed programmatically. A good example can be found in the file protocols/abinitio/util.cc. A SequenceAlignment and template PDB are converted into a set of NLGrouping's for use in the NonlocalAbinitio protocol.
Examples
Standard Topology
1a19a.flags (flags file)
-database ~/Rosetta/database
# target-related options
-frag3 1a19a/aa1a19a03_05.200_v1_3
-frag9 1a19a/aa1a19a09_05.200_v1_3
-fasta 1a19a/1a19a.fasta
-in:file:native 1a19a/1a19a.pdb
# non-local abinitio options
-abinitio:nonlocal_moves 1a19a/1a19a.pairings
-abinitio:nonlocal_close_chainbreaks
# chainbreak-related options
-jumps:ramp_chainbreaks
-jumps:overlap_chainbreak
-jumps:increase_chainbreak 0.5
# sampling options
-abinitio:skip_convergence_check
-abinitio:increase_cycles 10
-abinitio:rg_reweight 0.25
# output options
-nstruct 1000
-out:overwrite
-out:file:silent silent.out
1a19a.pairings (pairings file)
{
2,-110.466,126.421,174.600,97.539,49.731,-2.920
3,-119.756,145.382,-177.458,96.634,51.368,0.388
4,-132.235,119.796,176.520,95.461,49.639,3.564
5,-117.492,124.130,177.571,93.691,51.271,6.506
6,-107.749,99.134,179.550,92.793,49.314,9.639
49,-143.132,128.827,176.942,94.852,53.324,-6.631
50,-118.006,112.700,-171.815,93.025,50.630,-4.667
51,-105.538,117.641,178.295,92.132,51.602,-1.106
52,-102.343,117.860,179.175,91.170,48.933,1.425
53,-107.322,91.600,179.354,89.445,50.179,4.575
54,-71.162,135.072,176.466,89.341,47.329,7.090
84,-118.247,139.397,171.264,90.253,54.853,-7.446
85,-118.683,127.693,-173.555,88.952,51.548,-6.102
86,-116.318,111.870,175.979,87.492,51.458,-2.596
87,-100.612,112.101,-179.332,87.116,48.184,-0.701
88,-93.093,77.996,-179.257,85.043,48.589,2.460
}
command line:
nonlocal_abinitio.linuxgccrelease @1a19a.flags
Star Topology
Mostly used in comparative modeling. Same pairings file as above.
2kwba.flags (flags file)
-database ~/Rosetta/database
# target-related options
-frag3 2kwba/cs_frags.3mers
-frag9 2kwba/cs_frags.9mers
-fasta 2kwba/2kwba.fasta
-in:file:native 2kwba/2kwba.pdb
# non-local abinitio options
-abinitio:nonlocal_moves 2kwba/2kwba.pairings
-abinitio:nonlocal_close_chainbreaks
-abinitio:nonlocal_builder star
-abinitio:nonlocal_randomize_missing
# chainbreak-related options
-jumps:ramp_chainbreaks
-jumps:overlap_chainbreak
-jumps:increase_chainbreak 0.5
# sampling options
-abinitio:skip_convergence_check
-abinitio:increase_cycles 10
-abinitio:rg_reweight 0.25
# output options
-nstruct 1000
-out:overwrite
-out:file:silent silent.out
See Also
-
Structure prediction applications: A list of other applications to be used for structure prediction
- Abinitio-relax: Application for predicting protein structures using only sequence information
-
Membrane abinitio: Ab initio for membrane proteins.
-
Comparative modeling: Build structural models of proteins using one or more known structures as templates for modeling (uses the minirosetta application).
- Minirosetta: More information on the minirosetta application.
-
Metalloprotein ab initio: Ab inito modeling of metalloproteins.
-
Backrub: Create backbone ensembles using small, local backbone changes.
-
Comparative modeling: Build structural models of proteins using one or more known structures as templates for modeling.
- Floppy tail: Predict structures of long, flexible N-terminal or C-terminal regions.
-
Fold-and-dock: Predict 3-dimensional structures of symmetric homooligomers.
-
Molecular replacement protocols: Use Rosetta to build models for use in X-ray crystallography molecular replacement.
-
Prepare template for MR: Setup script for molecular replacement protocols.
-
Prepare template for MR: Setup script for molecular replacement protocols.
- Relax: "Locally" optimize structures, including assigning sidechain positions.
- Application Documentation: List of Rosetta applications
- Running Rosetta with options: Instructions for running Rosetta executables.
- Comparing structures: Essay on comparing structures
- Analyzing Results: Tips for analyzing results generated using Rosetta
- Solving a Biological Problem: Guide to approaching biological problems using Rosetta
- Commands collection: A list of example command lines for running Rosetta executable files