Directory dependency graph for task_operations:
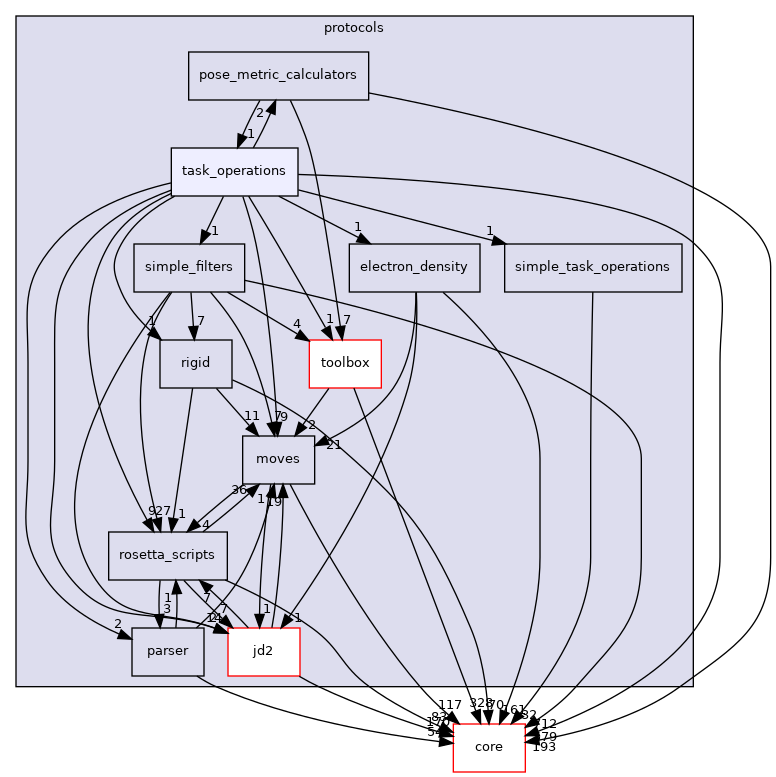
Files | |
| file | AlignedThreadOperation.cc |
| file | AlignedThreadOperation.fwd.hh |
| file | AlignedThreadOperation.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | AlignedThreadOperationCreator.hh |
| file | ConservativeDesignOperation.cc |
| file | ConservativeDesignOperation.fwd.hh |
| file | ConservativeDesignOperation.hh |
| file | ConservativeDesignOperationCreator.hh |
| file | CrystalContactsOperation.cc |
| Exclude crystal contacts from design. | |
| file | CrystalContactsOperation.fwd.hh |
| Exclude crystal contacts from design. | |
| file | CrystalContactsOperation.hh |
| Exclude crystal contacts from design. | |
| file | CrystalContactsOperationCreator.hh |
| Exclude crystal contacts from design. | |
| file | DatabaseThread.cc |
| picks a sequence from database by start and end position on the pose | |
| file | DatabaseThread.fwd.hh |
| file | DatabaseThread.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | DatabaseThreadCreator.hh |
| file | DesignAroundOperation.cc |
| file | DesignAroundOperation.fwd.hh |
| file | DesignAroundOperation.hh |
| file | DesignAroundOperationCreator.hh |
| file | DsspDesignOperation.cc |
| Design residues with selected amino acids depending on DSSP secondary structure assignment. All functionality here is included in the LayerDesign task operation, but this filter has significantly reduced overhead by avoiding slow SASA calculations. | |
| file | DsspDesignOperation.fwd.hh |
| file | DsspDesignOperation.hh |
| Design residues with selected amino acids depending on DSSP secondary structure assignment. All functionality here is included in the LayerDesign task operation, but this filter has significantly reduced overhead by avoiding slow SASA calculations. | |
| file | DsspDesignOperationCreator.hh |
| file | ImportUnboundRotamersOperation.cc |
| Import unbound (or native) rotamers into Rosetta! | |
| file | ImportUnboundRotamersOperation.fwd.hh |
| file | ImportUnboundRotamersOperation.hh |
| rotamer set operation forward declaration | |
| file | ImportUnboundRotamersOperationCreator.hh |
| file | InteractingRotamerExplosion.cc |
| file | InteractingRotamerExplosion.fwd.hh |
| file | InteractingRotamerExplosion.hh |
| TaskOperation class that oversamples rotamers interacting with a certain target. | |
| file | InteractingRotamerExplosionCreator.hh |
| file | JointSequenceOperation.cc |
| set designable residues to those observed in a set of structures | |
| file | JointSequenceOperation.fwd.hh |
| file | JointSequenceOperation.hh |
| set every position to be designable to residues observed in a set of structures | |
| file | JointSequenceOperationCreator.hh |
| file | LimitAromaChi2Operation.cc |
| eliminate aromatic rotamers, of which chi2 are around 0, 180 degree. @detail Chi2=0, 180 rotamers of aromatic residues ( PHE, TYR, HIS ) are not observed in nature very much, however Rosetta really like them. This is really pathology. For design purpose, we don't need them actually. | |
| file | LimitAromaChi2Operation.fwd.hh |
| file | LimitAromaChi2Operation.hh |
| rotamer set operation forward declaration | |
| file | LimitAromaChi2OperationCreator.hh |
| file | LinkResidues.cc |
| file | LinkResidues.fwd.hh |
| file | LinkResidues.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | LinkResiduesCreator.hh |
| file | ModifyAnnealer.cc |
| Task operation to set high and low temps for annealer as well as whether or not to do a quench step. | |
| file | ModifyAnnealer.fwd.hh |
| Task operation to set high and low temps for annealer as well as whether or not to do a quench step. | |
| file | ModifyAnnealer.hh |
| Task operation to set high and low temps for annealer as well as whether or not to do a quench step. | |
| file | ModifyAnnealerCreator.hh |
| file | MutationSetDesignOperation.cc |
| file | MutationSetDesignOperation.fwd.hh |
| file | MutationSetDesignOperation.hh |
| Sample a set of mutations each time packer is generated. | |
| file | pHVariantTaskOperation.cc |
| A TaskOp that enables sampling of protonation variants during repacking. The ionizable sidechains considered are ASP, GLU, LYS, TYR, and HIS. All other posiitons are restricted to repacking. The -pH_mode commandline option is still currently required to initially read the variants into the database. | |
| file | pHVariantTaskOperation.fwd.hh |
| During repacking, allow interchangeability of protonated and deprotonated variants. | |
| file | pHVariantTaskOperation.hh |
| file | pHVariantTaskOperationCreator.hh |
| file | PreventChainFromRepackingOperation.cc |
| file | PreventChainFromRepackingOperation.fwd.hh |
| file | PreventChainFromRepackingOperation.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | PreventChainFromRepackingOperationCreator.hh |
| file | PreventResiduesFromRepackingOperation.cc |
| file | PreventResiduesFromRepackingOperation.fwd.hh |
| file | PreventResiduesFromRepackingOperation.hh |
| TaskOperation class that prevents a vector of residues indices from repacking parsed as a string that is comma delimited ",". | |
| file | PreventResiduesFromRepackingOperationCreator.hh |
| file | ProteinCoreResFilter.cc |
| file | ProteinCoreResFilter.hh |
| file | ProteinCoreResFilterCreator.hh |
| file | ProteinInterfaceDesignOperation.cc |
| file | ProteinInterfaceDesignOperation.fwd.hh |
| file | ProteinInterfaceDesignOperation.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | ProteinInterfaceDesignOperationCreator.hh |
| file | PruneBuriedUnsatsOperation.cc |
| Eliminate rotamers that contain a sidechain buried unsat and make no other sidechain h-bonds @detail This is intended to speed up packing by eliminating useless polar rotamers. This can also help to reduce the number of buried unsats in designs because Rosetta can't pack them. | |
| file | PruneBuriedUnsatsOperation.fwd.hh |
| Forward declaration for PruneBuriedUnsatsOperation. | |
| file | PruneBuriedUnsatsOperation.hh |
| Eliminate rotamers that contain a sidechain buried unsat and make no other sidechain h-bonds @detail This is intended to speed up packing by eliminating useless polar rotamers. This can also help to reduce the number of buried unsats in designs because Rosetta can't pack them. | |
| file | PruneBuriedUnsatsOperationCreator.hh |
| Creator for PruneBuriedUnsatsOperation. | |
| file | ReadResfileFromDB.cc |
| file | ReadResfileFromDB.fwd.hh |
| Forward declaration of the ReadResfileFromDB class. | |
| file | ReadResfileFromDB.hh |
| read a refile indexed by the input structure tag from a supplied relational database | |
| file | ReadResfileFromDBCreator.hh |
| file | ResfileCommandOperation.cc |
| Applies the equivalent of a resfile line (without the resnums) to residues specified in a residue selector. | |
| file | ResfileCommandOperation.fwd.hh |
| Applies the equivalent of a resfile line (without the resnums) to residues specified in a residue selector. | |
| file | ResfileCommandOperation.hh |
| file | ResfileCommandOperationCreator.hh |
| file | ResidueProbDesignOperation.cc |
| file | ResidueProbDesignOperation.fwd.hh |
| file | ResidueProbDesignOperation.hh |
| Uses a probability distribution to sample allowed residues at a position, adding those types to the packer - only if they are designable. | |
| file | ResidueProbDesignOperationCreator.hh |
| file | RestrictAAsFromProbabilities.cc |
| A class to restrict designable amino acids depending on the probabilities from a PerResidueProbabilitiesMetric. | |
| file | RestrictAAsFromProbabilities.fwd.hh |
| A class to restrict designable amino acids depending on the probabilities from a PerResidueProbabilitiesMetric. | |
| file | RestrictAAsFromProbabilities.hh |
| file | RestrictAAsFromProbabilitiesCreator.hh |
| file | RestrictByCalculatorsOperation.cc |
| A class that applies arbitrary calculators (whose calculations return std::set< core::Size >) to restrict a PackerTask. | |
| file | RestrictByCalculatorsOperation.fwd.hh |
| A class that applies arbitrary calculators (whose calculations return std::set< core::Size >) to restrict a PackerTask. | |
| file | RestrictByCalculatorsOperation.hh |
| A class that applies arbitrary calculators (whose calculations return std::set< core::Size >) to restrict a PackerTask. | |
| file | RestrictByCalculatorsOperationCreator.hh |
| file | RestrictChainToRepackingOperation.cc |
| file | RestrictChainToRepackingOperation.fwd.hh |
| file | RestrictChainToRepackingOperation.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | RestrictChainToRepackingOperationCreator.hh |
| file | RestrictIdentitiesAtAlignedPositions.cc |
| file | RestrictIdentitiesAtAlignedPositions.fwd.hh |
| file | RestrictIdentitiesAtAlignedPositions.hh |
| file | RestrictIdentitiesAtAlignedPositionsCreator.hh |
| file | RestrictIdentitiesOperation.cc |
| Restricts specified residue types to only repack, no design. | |
| file | RestrictIdentitiesOperation.fwd.hh |
| file | RestrictIdentitiesOperation.hh |
| file | RestrictIdentitiesOperationCreator.hh |
| file | RestrictInterGroupVectorOperation.cc |
| file | RestrictInterGroupVectorOperation.fwd.hh |
| file | RestrictInterGroupVectorOperation.hh |
| restricts design to only those residues between two groups of structures | |
| file | RestrictInterGroupVectorOperationCreator.hh |
| file | RestrictNativeResiduesOperation.cc |
| Restrict every residue in the current pose that is native to repacking. ie, only allow mutated positions to be designed. | |
| file | RestrictNativeResiduesOperation.fwd.hh |
| file | RestrictNativeResiduesOperation.hh |
| Restrict every residue in the current pose that is native to repacking. ie, only allow mutated positions to be designed. | |
| file | RestrictNativeResiduesOperationCreator.hh |
| file | RestrictNonSurfaceToRepackingOperation.cc |
| file | RestrictNonSurfaceToRepackingOperation.fwd.hh |
| file | RestrictNonSurfaceToRepackingOperation.hh |
| file | RestrictNonSurfaceToRepackingOperationCreator.hh |
| file | RestrictOperationsBase.cc |
| Base class for PoseMetricCalculator-using TaskOperations. | |
| file | RestrictOperationsBase.fwd.hh |
| Base class for PoseMetricCalculator-using TaskOperations. | |
| file | RestrictOperationsBase.hh |
| Base class for PoseMetricCalculator-using TaskOperations. | |
| file | RestrictResiduesToRepackingOperation.cc |
| file | RestrictResiduesToRepackingOperation.fwd.hh |
| file | RestrictResiduesToRepackingOperation.hh |
| TaskOperation class that restricts a vector of core::Size defined residues to repacking when parsed, it takes in a string and splits by ",". | |
| file | RestrictResiduesToRepackingOperationCreator.hh |
| file | RestrictToAlignedSegments.cc |
| file | RestrictToAlignedSegments.fwd.hh |
| file | RestrictToAlignedSegments.hh |
| file | RestrictToAlignedSegmentsCreator.hh |
| file | RestrictToCDRH3Loop.cc |
| file | RestrictToCDRH3Loop.fwd.hh |
| Forward declaration of the RestrictToCDRH3Loop class. | |
| file | RestrictToCDRH3Loop.hh |
| This class allows the selection for packing of the Antibody CDR-H3 loop by taking advantage of the PDB numbering schemes that are commonly used for Antibodies. | |
| file | RestrictToCDRH3LoopCreator.hh |
| file | RestrictToInterfaceOperation.cc |
| TaskOperation class that finds an interface and leaves it mobile in the PackerTask. | |
| file | RestrictToInterfaceOperation.fwd.hh |
| Forward declaration of a TaskOperation class that finds an interface leaves it mobile in the PackerTask. | |
| file | RestrictToInterfaceOperation.hh |
| TaskOperation class that finds an interface and makes it mobile in the PackerTask. | |
| file | RestrictToInterfaceOperationCreator.hh |
| file | RestrictToInterfaceVectorOperation.cc |
| TaskOperation class that finds an interface based on: core/select/util/interface_vector_calculate.hh and leaves it mobile in the PackerTask. | |
| file | RestrictToInterfaceVectorOperation.fwd.hh |
| Forward declaration of a TaskOperation class that finds an interface leaves it mobile in the PackerTask. | |
| file | RestrictToInterfaceVectorOperation.hh |
| TaskOperation class that finds an interface based on InterfaceVectorDefinition and leaves it mobile in the PackerTask. Serves mostly to wrap InterfaceVectorDefinition into a TaskOperation. see src/core/select/util/interface_vector_calculate.hh. | |
| file | RestrictToInterfaceVectorOperationCreator.hh |
| file | RestrictToMoveMapChiOperation.cc |
| file | RestrictToMoveMapChiOperation.fwd.hh |
| file | RestrictToMoveMapChiOperation.hh |
| Makes it easier to use TF and MM in the same class. | |
| file | RestrictToMoveMapChiOperationCreator.hh |
| file | RestrictToNeighborhoodOperation.cc |
| TaskOperation class that finds a neighborhood and leaves it mobile in the PackerTask. | |
| file | RestrictToNeighborhoodOperation.fwd.hh |
| Forward declaration of a TaskOperation class that finds a neighborhood; leaves it mobile in the PackerTask. | |
| file | RestrictToNeighborhoodOperation.hh |
| TaskOperation class that finds a neighborhood and makes it mobile in the PackerTask. | |
| file | RestrictToNeighborhoodOperationCreator.hh |
| file | RestrictToTerminiOperation.cc |
| file | RestrictToTerminiOperation.fwd.hh |
| Restrict to packing only the residues at either or both termini. | |
| file | RestrictToTerminiOperation.hh |
| file | RestrictToTerminiOperationCreator.hh |
| file | RetrieveStoredTaskOperation.cc |
| Retrieves a stored task from the pose's CacheableData. Must be used in conjunction with the StoredTaskMover. Allows storage/retrieval of a task so that particular sets of residues can be stably addressed throughout the entirety of a RosettaScripts protocol. | |
| file | RetrieveStoredTaskOperation.fwd.hh |
| file | RetrieveStoredTaskOperation.hh |
| Retrieves a stored task from the pose's CacheableData. Must be used in conjunction with the StoredTaskMover. Allows storage/retrieval of a task so that particular sets of residues can be stably addressed throughout the entirety of a RosettaScripts protocol. | |
| file | RetrieveStoredTaskOperationCreator.hh |
| file | SelectByDeltaScoreOperation.cc |
| file | SelectByDeltaScoreOperation.fwd.hh |
| file | SelectByDeltaScoreOperation.hh |
| file | SelectByDeltaScoreOperationCreator.hh |
| file | SelectByDensityFitOperation.cc |
| file | SelectByDensityFitOperation.fwd.hh |
| file | SelectByDensityFitOperation.hh |
| file | SelectByDensityFitOperationCreator.hh |
| file | SelectBySASAOperation.cc |
| file | SelectBySASAOperation.fwd.hh |
| file | SelectBySASAOperation.hh |
| file | SelectBySASAOperationCreator.hh |
| file | SelectResiduesWithinChain.cc |
| file | SelectResiduesWithinChain.fwd.hh |
| file | SelectResiduesWithinChain.hh |
| file | SelectResiduesWithinChainCreator.hh |
| file | SeqprofConsensusOperation.cc |
| set designable residues to those observed in sequence profile | |
| file | SeqprofConsensusOperation.fwd.hh |
| file | SeqprofConsensusOperation.hh |
| file | SeqprofConsensusOperationCreator.hh |
| file | SequenceMotifTaskOperation.cc |
| A TaskOp that takes a regex-like pattern and turns it into a set of design residues. The string should identify what to do for each position. An X indicates any residue, and is the same as [A_Z]. Anything other than a charactor should be in []. The ^ denotes a not. An example Regex for glycosylation is NX[ST]. This would design an ASN into the first position, skip the second, and allow S and T mutations only at the third position. N[^P][ST] would denote that at the second position, we do not allow proline. Sets of charactors using _ can be denoted, even though this doesnt really help us in design. In the future one can imagine having sets of polars, etc. etc. This TaskOp is like a simple RESFILE in a string. | |
| file | SequenceMotifTaskOperation.fwd.hh |
| A TaskOp that takes a regex-like pattern and turns it into a set of design residues. The string should identify what to do for each position. An X indicates any residue, and is the same as [A_Z]. Anything other than a charactor should be in []. The ^ denotes a not. An example Regex for glycosylation is NX[ST]. This would design an ASN into the first position, skip the second, and allow S and T mutations only at the third position. N[^P][ST] would denote that at the second position, we do not allow proline. Sets of charactors using _ can be denoted, even though this doesnt really help us in design. In the future one can imagine having sets of polars, etc. etc. This TaskOp is like a simple RESFILE in a string. | |
| file | SequenceMotifTaskOperation.hh |
| file | SequenceMotifTaskOperationCreator.hh |
| file | SetIGTypeOperation.cc |
| Task operation to set interaction graph type (linmem, lazy or double lazy) | |
| file | SetIGTypeOperation.fwd.hh |
| Task operation to set Interaction graph Type. | |
| file | SetIGTypeOperation.hh |
| Task operation to set interaction graph type (linear memory, lazy or double lazy) | |
| file | SetIGTypeOperationCreator.hh |
| file | STMStoredTask.cc |
| CacheableData wrapper for PackerTask storage. | |
| file | STMStoredTask.fwd.hh |
| file | STMStoredTask.hh |
| file | StoreCombinedStoredTasksMover.cc |
| The StoreCombinedStoredTasksMover HAS BEEN DEPRECATED. It was used to combine several STMStoredTasks (protocols/task_operations/STMStoredTask.cc), but has been replaced by the StoreCompoundTaskMover (protocols/task_operations/StoreCompoundTaskMover.cc), which is much more flexible. It is being moved out of devel/matdes/ into protocols/task_operations/ to support the publication King et al., Nature 2014. | |
| file | StoreCombinedStoredTasksMover.fwd.hh |
| file | StoreCombinedStoredTasksMover.hh |
| Headers for StoreCombinedStoredTasksMover class – DEPRECATED (see .cc file) | |
| file | StoreCombinedStoredTasksMoverCreator.hh |
| file | StoreCompoundTaskMover.cc |
| Combine tasks using boolean logic for residues that are packable or designable, assign new packing behavior to residues that match or do not match the specified criteria, and store the resulting task in the pose's cacheable data. TODO (Jacob): Add option to combine allowed amino acid sets. | |
| file | StoreCompoundTaskMover.fwd.hh |
| file | StoreCompoundTaskMover.hh |
| file | StoreCompoundTaskMoverCreator.hh |
| file | StoreTaskMover.cc |
| The StoreTaskMover allows you to create a PackerTask at some point during a RosettaScripts run and save it for access later during the same run. Can be useful for mutating/analyzing a particular set of residues using many different movers/filters. | |
| file | StoreTaskMover.fwd.hh |
| file | StoreTaskMover.hh |
| Headers for StoreTaskMover class. | |
| file | StoreTaskMoverCreator.hh |
| This class will create instances of Mover StoreTaskMover for the MoverFactory. | |
| file | ThreadSequenceOperation.cc |
| file | ThreadSequenceOperation.fwd.hh |
| file | ThreadSequenceOperation.hh |
| TaskOperation class that restricts a chain to repacking. | |
| file | ThreadSequenceOperationCreator.hh |